

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PKNH_0205600 | OTHER | 0.925805 | 0.061467 | 0.012728 |
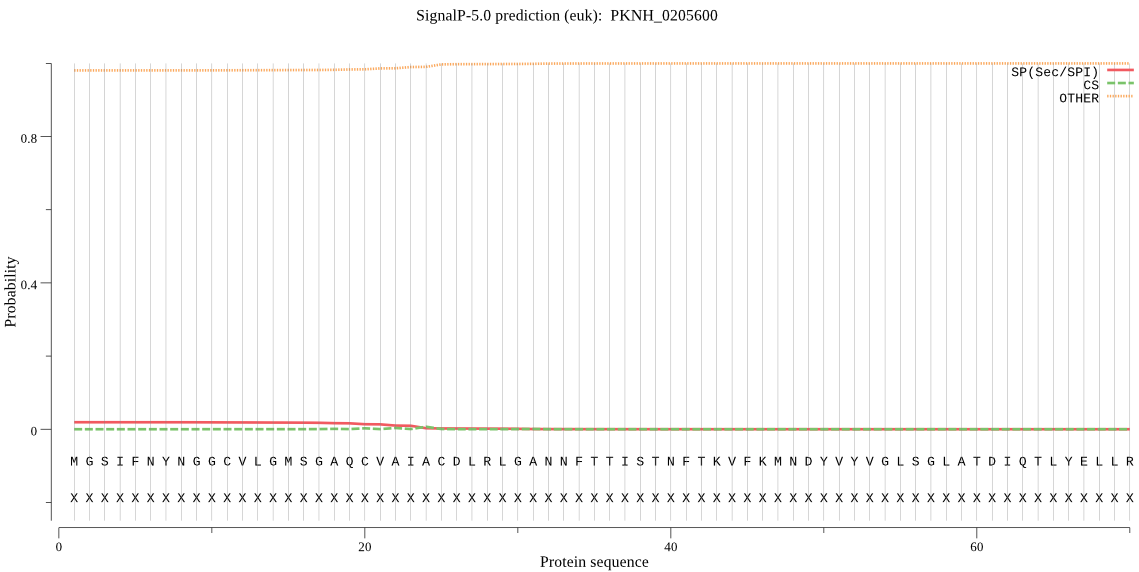
MGSIFNYNGGCVLGMSGAQCVAIACDLRLGANNFTTISTNFTKVFKMNDYVYVGLSGLAT DIQTLYELLRYRVNLYEIRQETPMDIDCFANMLSSILYANRFSPYFVNPIVVGFRVKHTF DDEGKKIRTFEPYLTAYDLIGAKCETKDFVVNGVTSEQLYGMCESLYIKDQDKDGLFETI SQCLLSALDRDCLSGWGAEVYVLTPDSIMKKKLKARMD