

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
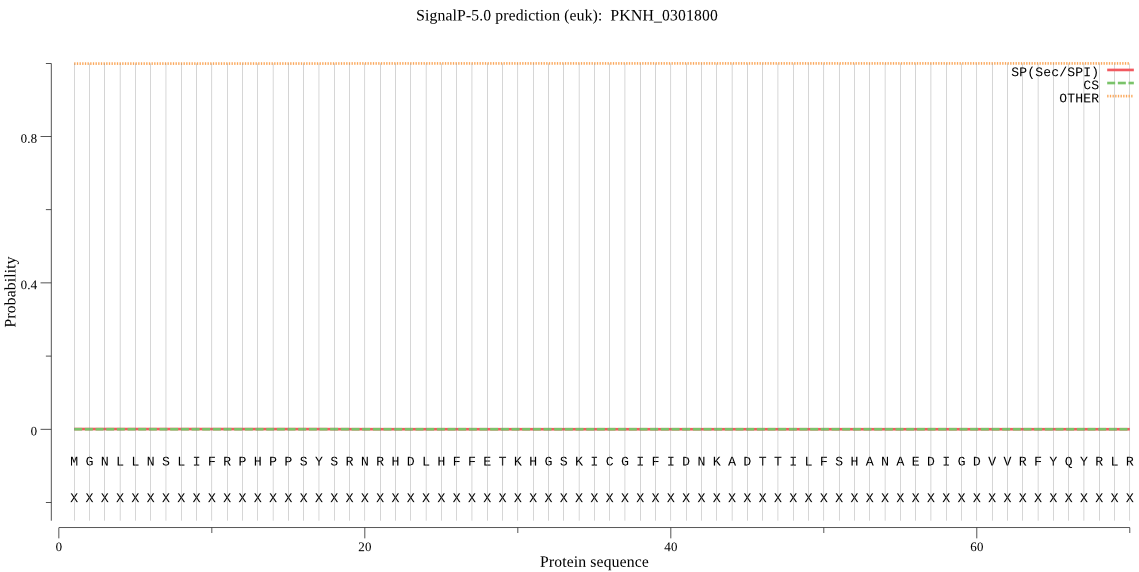
| PKNH_0301800 | OTHER | 0.997189 | 0.000673 | 0.002138 |
MGNLLNSLIFRPHPPSYSRNRHDLHFFETKHGSKICGIFIDNKADTTILFSHANAEDIGD VVRFYQYRLRRLGLNLFAYDYSGYGHSSGHPTEAHVYNDVEAAYDYLVKVLRVPRHSIIA YGRSLGSAASVHIATKKNLLGLILQAPLASIHRVKLKLKFTLPYDSFCNIDKVHMINCPI LFIHGTKDKLLSYHGTEEMIRRTNVNTYFMFIEGGGHNDLDSSYGNQVYAALVAFLYVLK NNIRENVNSVYDISNVNMMKLRNMFISNNTKNMKERVKEKKRKGEENGMVSGSAGSVNGR QGNRNESRGSNEVLNSFMSNMERWYTDESIFRVYTKDSGYDSSQSLSSVLGSAESTMRHI PDNTFYESDTNISIEDLCRLAGGKNNLNRSKLYNGKDSGASCSSFTKRGWIDRSDGAVGN HRNGHRGVSRERYPDGRRIDSKDGRDRSRMKVCSSYVYNPARRENISPSNMDHSSSQCSR SDVYMGSEQDLAQRGNGTCMGGSSSEKVNSRRSGESRNIYSVDRVPRISQDMKSVGSSPV SERGSYARSYEQCNNGREETTIRGHRNGEQYSSESRKGGGNMNPSKASLASVSSNDSKIS SVGRGPFKEERAKVMNESRRNMQSDTIKQGSSVVKEGCATSEHVYSKNYYKKEGNRIRKE MENKITDINNQKINRYNEFIYGNQENKMGDCPNEIMEPVQREVSSASSSTLNNLNSLFAY
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PKNH_0301800 | 249 S | ENVNSVYDI | 0.992 | unsp | PKNH_0301800 | 249 S | ENVNSVYDI | 0.992 | unsp | PKNH_0301800 | 249 S | ENVNSVYDI | 0.992 | unsp | PKNH_0301800 | 307 S | NRNESRGSN | 0.995 | unsp | PKNH_0301800 | 338 S | YTKDSGYDS | 0.995 | unsp | PKNH_0301800 | 373 S | DTNISIEDL | 0.996 | unsp | PKNH_0301800 | 429 S | HRGVSRERY | 0.997 | unsp | PKNH_0301800 | 467 S | RENISPSNM | 0.994 | unsp | PKNH_0301800 | 510 S | EKVNSRRSG | 0.994 | unsp | PKNH_0301800 | 513 S | NSRRSGESR | 0.993 | unsp | PKNH_0301800 | 538 S | SVGSSPVSE | 0.992 | unsp | PKNH_0301800 | 541 S | SSPVSERGS | 0.995 | unsp | PKNH_0301800 | 575 S | YSSESRKGG | 0.992 | unsp | PKNH_0301800 | 16 S | PHPPSYSRN | 0.994 | unsp | PKNH_0301800 | 117 S | VPRHSIIAY | 0.993 | unsp |