

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
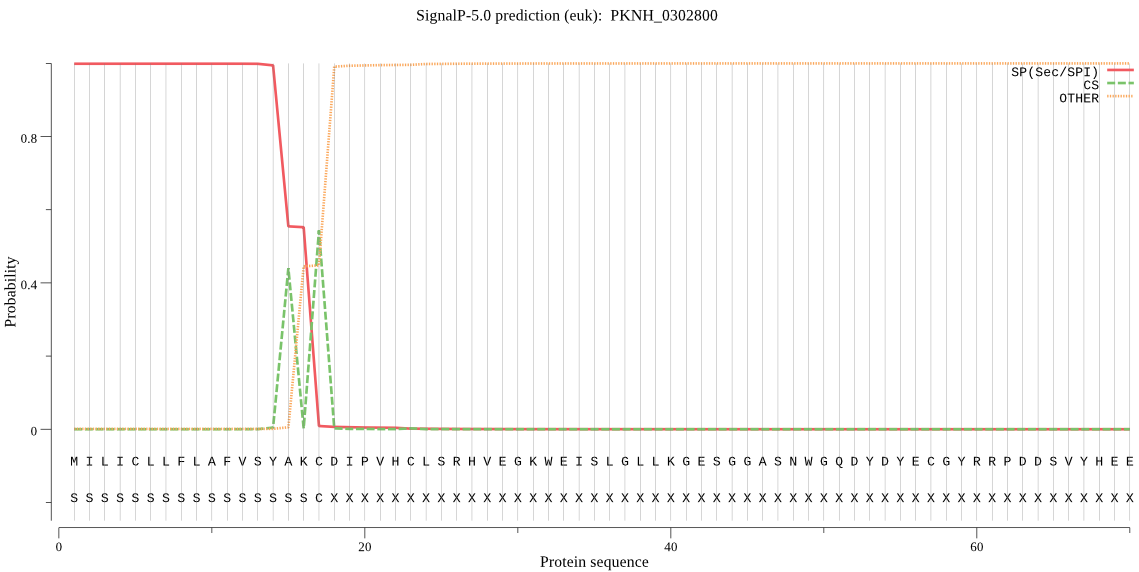
| PKNH_0302800 | SP | 0.000681 | 0.999317 | 0.000002 | CS pos: 17-18. AKC-DI. Pr: 0.8562 |
MILICLLFLAFVSYAKCDIPVHCLSRHVEGKWEISLGLLKGESGGASNWGQDYDYECGYR RPDDSVYHEELDPDHLKGRFEEKKKQIIQFNTDRSINIIENGEINPEYSGFWRIVYDEGL YMEVYKQNDEKEIYFSFFKFQRRNDASYSYCNRLIMGVVNVYHLNGSEGSSSSSGVSGVD DLSDQNVRGKRGSKLKDVFLVKMEHNGKNGLTPKRINLSRERFHFVGSKEGAPKSSSHSI DYYNDNYINFDMFTMKRLCWYGKKVGTPDEPTNKIPVEIISPLVLDPDTHNKNYANLKLE WGGVNDKRIQRVNHGQPHTDKALTRLRTLPKHTEKSNKGMKQNSIFNTYREKDVSLAHFD WTDEADVRERLNGKWVKVIDEAIDQKECGSCYANSAALIINSRLRIKYNYIKNIELLSFS NKQLVLCDFFNQGCNGGYIYLSLKYAYENFLFTNKCFVRYSSLYLNKDTKNSALCDRFDT FKAFLKKGSGEDWSSAHVGPLHRQTKRVRQHEPSGGSEENVGGEDMFVGEEVDKSSTQGE EHSGGNKHSRHHMGDVDDVRDEQDLPKDHELSEGYLPVDDSVRDASEVDMLKLRECDAKV KVTKFEYLDIEDEEDLKKYLYHNGPVAAAIEPSKNFPAYKEGILTGKFIKMSDGGESNAY VWNKVDHAVVIVGWGEDTVENLKKKKKKVHHYSDDLDAYASGKEDKAGGKVVKYWKILNS WGTQWGYDGYFYILRDENYFSIKSYLLACDVSLFIKRGDSGVSQG
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PKNH_0302800 | 489 S | LKKGSGEDW | 0.996 | unsp | PKNH_0302800 | 489 S | LKKGSGEDW | 0.996 | unsp | PKNH_0302800 | 489 S | LKKGSGEDW | 0.996 | unsp | PKNH_0302800 | 581 S | PVDDSVRDA | 0.996 | unsp | PKNH_0302800 | 586 S | VRDASEVDM | 0.996 | unsp | PKNH_0302800 | 701 S | DAYASGKED | 0.995 | unsp | PKNH_0302800 | 760 S | KRGDSGVSQ | 0.994 | unsp | PKNH_0302800 | 65 S | RPDDSVYHE | 0.99 | unsp | PKNH_0302800 | 177 S | SSGVSGVDD | 0.995 | unsp |