

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
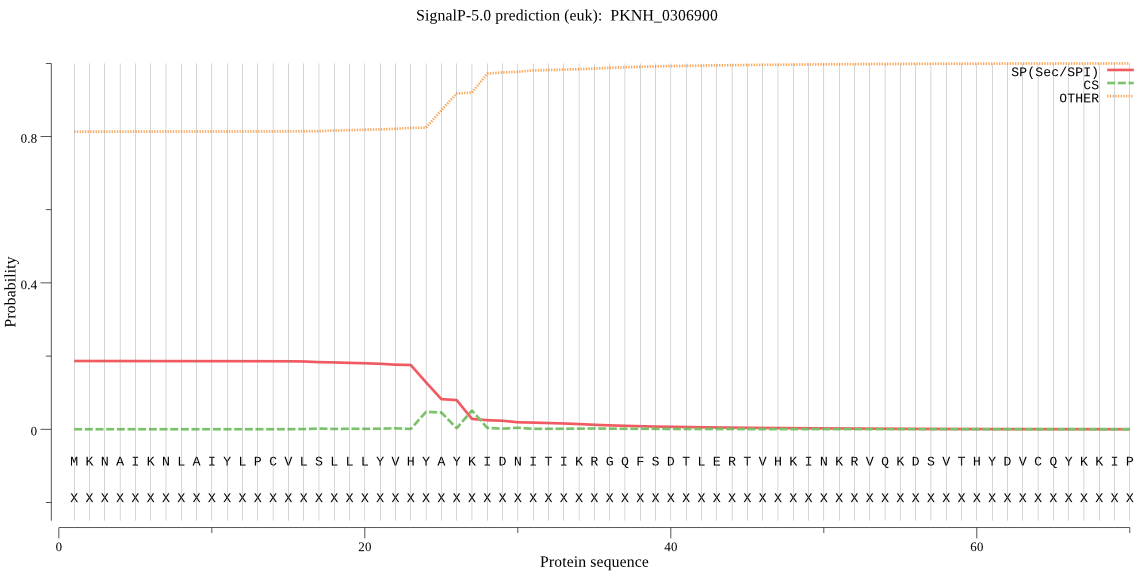
| PKNH_0306900 | SP | 0.488535 | 0.510000 | 0.001465 | CS pos: 25-26. HYA-YK. Pr: 0.4548 |
MKNAIKNLAIYLPCVLSLLLYVHYAYKIDNITIKRGQFSDTLERTVHKINKRVQKDSVTH YDVCQYKKIPKILTERNKFKDSQKVKYIVGIENTCDDTCICILDSNLKIVKNVIISHFKV VHEYEGVYPFFISSINQFFLKPYMEKTLEGIDERLICCFAFSACPGIAKSMEAAKNYIGE KKKQNGSIQVSPINHVFAHVLSPLFFHVYDDKNTYTNSGQYKEGKRDQVEKELCIDMHIS EEVRKDKTQMEKVKDILHILNSNDVTKQKMEFLSEDKFLNCGNYVLRGEMNKEAPQSVES QAQQNVAYTRKTTSSSHKTQYSEDAPQRSYLTDGYICALLSGGSTQIYKVQKNKKNDINL CKLSQTVDISIGDIIDKVARLLGLPVGLGGGPFLERKAEEFIKRMKEEHLSGEACADPFQ PFPVPFSANNRIDFSFSGIYNHVRKIIEELKKGESFEREKDRYAYFLQKNIFNHLLTQIN KIMYFSELHFNVKELFIVGGVGCNNFLITELKNMAMKRSQLPFQLNEFKKLKKRLSKKIK KISEKNILRSKVSKEVLKGEIEFSASLNWDVYLKFLLKRKTEQDILSALECFNFEDFSKL KEEGHFLLEGHSPFSKSEMPWSVHKTPLNLSRDNAAMICFSAFLNLHNKSHIYEDVSQIE IKPTVKTQVENNFLLFSDIIIFDVFLHQFGAQGG
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PKNH_0306900 | 455 S | KKGESFERE | 0.992 | unsp | PKNH_0306900 | 455 S | KKGESFERE | 0.992 | unsp | PKNH_0306900 | 455 S | KKGESFERE | 0.992 | unsp | PKNH_0306900 | 536 S | KKRLSKKIK | 0.995 | unsp | PKNH_0306900 | 543 S | IKKISEKNI | 0.993 | unsp | PKNH_0306900 | 615 S | HSPFSKSEM | 0.995 | unsp | PKNH_0306900 | 657 S | YEDVSQIEI | 0.995 | unsp | PKNH_0306900 | 297 S | EAPQSVESQ | 0.992 | unsp | PKNH_0306900 | 316 S | TTSSSHKTQ | 0.992 | unsp |