-
Computed_GO_Component_IDs:
-
Computed_GO_Components:
-
Computed_GO_Function_IDs:
-
Computed_GO_Functions:
-
Computed_GO_Process_IDs:
-
Computed_GO_Processes:
-
Curated_GO_Component_IDs:
-
Curated_GO_Components:
-
Curated_GO_Function_IDs:
-
Curated_GO_Functions:
-
Curated_GO_Processes:
No Results
-
Fasta :-
>PKNH_0314000
MFHHFRGRNALHNYLSRCACGKFPPGVIPYRNRWPKVRSLTVLETLRRPHSGYSRRGDPS
HTTKHRRDCKGGKADSFKEKHERDGWSERSEKINTEMSLYYPESNSFPFVTTGILLGMVG
ISSFFQFLIYTLKFCEDESSILLKVNEFLDHLDAYFIIHDFEGGPGEGHSTRGRDAVKSG
LFSIKYLTSPFFANENILQCAVQLSTFFLASRFLEKQFGSLKFGALFISASILSNLLTHN
FLKYLTNQVESLNSLNFVLIHPSGSMAFICALCSICFKNCAIWKDIPVHCSILVVPYLLS
SFYGLLSLYKIGKNTLENTHGEVAPSERNSLQSVDAKDHPYEDPPHLGMPHTTRSSHAND
ITNPLHSSDKEQPNAPPSNKHVQNETMEILRNFLIVKACDEVIKRRKKENMFPNKKIQNL
KNESLKNINEINNRSKKIFFGLSSSFTDICGILLASCGSFLFKMLR
- Download Fasta
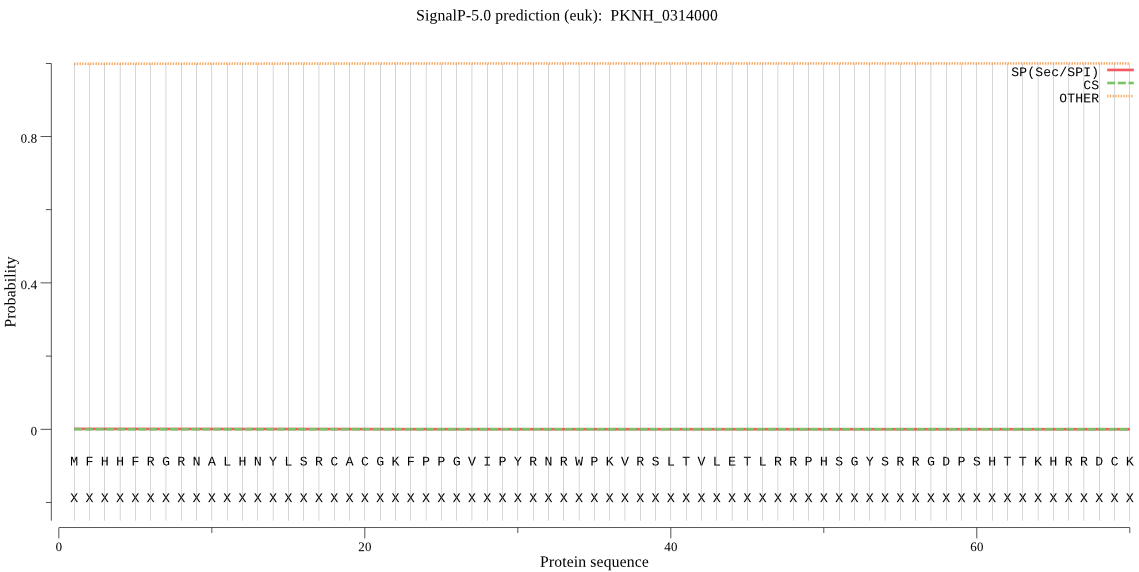
-
MitoProt II - v1.101
File : /home/rajan/sadaf/4480_mitoprot/test/PKNH_0314000.fa
Sequence name : PKNH_0314000
Sequence length : 466
VALUES OF COMPUTED PARAMETERS
Coef20 : 4.183
CoefTot : -2.113
ChDiff : 14
ZoneTo : 57
KR : 12
DE : 1
CleavSite : 58
HYDROPHOBIC SCALE USED
GES KD GVH1 ECS
H17 : 2.241 1.947 0.438 0.769
MesoH : 0.560 1.000 -0.043 0.408
MuHd_075 : 42.205 25.476 12.250 9.283
MuHd_095 : 39.061 20.274 8.282 8.233
MuHd_100 : 32.294 17.738 8.388 7.900
MuHd_105 : 38.506 19.937 8.342 9.490
Hmax_075 : 10.600 7.875 3.920 3.970
Hmax_095 : 6.100 10.700 -1.904 3.460
Hmax_100 : 6.100 11.100 2.120 3.760
Hmax_105 : 9.217 9.450 0.700 4.083
CLASS NOT-MITO MITO(/CHLORO)
DFM : 0.0112 0.9888
DFMC : 0.0235 0.9765
This protein is probably imported in mitochondria.
f(Ser) = 0.0702 f(Arg) = 0.1754 CMi = 0.22235
CMi is the Chloroplast/Mitochondria Index
It has been proposed by Von Heijne et al
(Eur J Biochem,1989, 180: 535-545)
-
Fasta :-
>PKNH_0314000
ATGTTCCACCATTTCAGGGGGAGGAATGCACTACATAACTACCTATCAAGATGCGCCTGC
GGGAAGTTCCCGCCTGGGGTTATCCCCTACAGGAACCGCTGGCCCAAAGTAAGAAGCCTA
ACTGTGTTGGAAACTCTGAGAAGACCCCACAGTGGGTACAGTAGAAGGGGCGACCCATCG
CACACCACAAAGCATAGAAGGGATTGCAAAGGTGGCAAAGCAGATTCCTTCAAAGAAAAA
CATGAAAGGGACGGATGGAGCGAACGCAGTGAAAAAATAAACACAGAAATGTCCCTTTAC
TATCCAGAGAGTAACTCCTTCCCATTTGTGACTACCGGCATCTTGTTGGGCATGGTGGGC
ATTTCGTCCTTTTTTCAATTTTTAATTTACACCCTGAAATTTTGTGAGGATGAGTCAAGC
ATCCTGCTAAAAGTAAACGAGTTCCTGGACCATCTGGATGCCTATTTTATAATTCACGAC
TTTGAGGGGGGCCCAGGTGAAGGACACAGTACAAGAGGACGTGACGCCGTGAAGTCTGGC
CTCTTCAGCATCAAATATTTGACGTCTCCATTTTTCGCCAATGAAAATATCCTACAATGC
GCAGTTCAGCTATCCACCTTCTTTTTGGCCTCGAGATTTCTTGAAAAACAATTTGGGTCC
TTAAAATTTGGCGCACTATTCATAAGCGCTTCCATACTTTCTAACCTTCTAACACATAAC
TTCTTAAAATACCTTACGAACCAAGTGGAATCATTAAACTCCCTTAACTTTGTTCTTATA
CACCCTTCTGGGTCCATGGCTTTCATTTGTGCCTTGTGCTCCATTTGCTTCAAAAATTGC
GCCATCTGGAAGGATATCCCAGTCCACTGCTCCATTTTAGTCGTTCCTTATCTTCTTTCT
TCCTTCTACGGTCTATTATCTTTGTACAAAATAGGAAAGAATACTTTGGAAAATACTCAC
GGGGAGGTTGCTCCCAGTGAGAGGAACTCCCTCCAAAGTGTAGATGCTAAAGACCATCCC
TATGAAGATCCCCCACATTTGGGTATGCCCCACACAACAAGGAGCAGCCATGCCAACGAC
ATAACTAACCCCCTCCACAGCAGTGACAAAGAACAACCAAATGCCCCCCCCAGTAATAAA
CACGTCCAAAATGAAACCATGGAAATTTTAAGGAACTTCCTAATCGTAAAGGCATGTGAT
GAAGTTATTAAGAGGCGCAAAAAGGAAAATATGTTTCCAAACAAAAAGATACAGAACCTC
AAAAATGAATCGCTTAAAAATATCAACGAAATTAATAACAGGTCGAAGAAAATATTTTTT
GGCTTGTCTTCTTCCTTTACTGACATATGTGGCATCCTGCTTGCATCATGCGGGTCTTTC
CTTTTTAAAATGCTCAGGTAG
- Download Fasta
|
ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method |
|
PKNH_0314000 | 326 S | EVAPSERNS | 0.991 | unsp | PKNH_0314000 | 326 S | EVAPSERNS | 0.991 | unsp | PKNH_0314000 | 326 S | EVAPSERNS | 0.991 | unsp | PKNH_0314000 | 51 S | RRPHSGYSR | 0.992 | unsp | PKNH_0314000 | 76 S | GKADSFKEK | 0.995 | unsp |


