

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PKNH_0413200 | SP | 0.000158 | 0.999841 | 0.000000 | CS pos: 22-23. ARC-AP. Pr: 0.5842 |
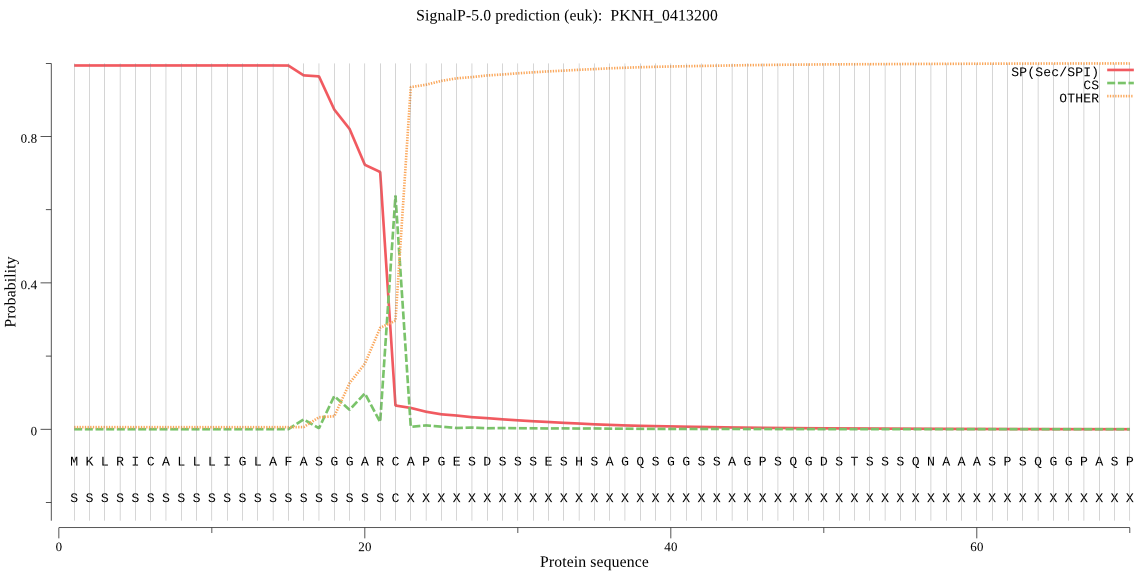
MKLRICALLLIGLAFASGGARCAPGESDSSSESHSAGQSGGSSAGPSQGDSTSSSQNAAA SPSQGGPASPPTDNPESSVGDNPASLSQGTGNGEVSSADSTSGTPGAEGDNAATVQSPEV TNTEVSPQPEANSPDTPPVSQPSSVPTSSSGDSPVPATSSASFTNPIQVKASLLKNYKGV KVTGPCGSYFQVYLVPYLYMNVNSSNSEIEMEPLFMKVDNKIKFEKEKHLLHNICANNKT FKLVLYVYEGVLTIKWKVYPAEGGEGSDKKVDIRKYKMKDIGQPITSIQVMMVTERDGTI YVESKNFSIMNDIPEKCDAIANQCFMSGVLDIQKCYHCTLLLQEKEKAEECFKFVSTEVK NRFEEIQTKGEDEKNPNEAELEETIDNIFTKIYGGEENLHKEVNQLAILDYSLKYELLKY CLLMKEVDASGTLDNQELPNPEDVFAHITTLLQNNNELNVFSLKNKMKNPALCLKEVDQW IGSKTGLTLPALEHSTVQNNEETFDESEDDISGSTTTEGADLYPLHFSDKLFCNYDYCDR TKDTSSCLSKMEVHDQGNCATSWLFASKLHLETIKCMKGYKHIPSSALYVANCSNKETHN KDKCHTPSNPLEFLHTLEETKFLPAESDLPYCYKKVGNDICPEPKSHWQNLWTNIKLVDA QYQPNSISTKGYTEYQSKYFKGNMDAFIKLVKSEIMNKGSVIAYVKATGALSFDLNGKKV QNLCGEEGTPDLAVNIIGYGNYINEEGIKKSYWLLQNSWGYYWGNEGTFKVDMHGPDDCE HNFIHTAAVFNLDIPVVIPAPNGDPEINSYYLKNSPDFYKNLYYNALDGECGSTPCHSVE GQDTTVEQTTGQEASREDTEEKATEQVGASGAGVATVATTGTGAAPGTGAKAGAEAGAGA ETEATKATELQEPKLQQEQEQQTQSPQQPPQQGLTNENAQQADGEVVQGAPGQAVATHPV PSGEGNGTGGETVPGNTNSVQSTPQNSGSSAEIGQTNASTNGAEPPVSDPVETPSPVELS KLTGTTGINVDGVTRVLHFLKNVKNGKVTSSFVTYDNESAIGDKSCSRVQSSDLDKLDDC VKFCEENWDACKGSVSPGYCLTKKRGNNECFFCFV
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PKNH_0413200 | 78 S | NPESSVGDN | 0.997 | unsp | PKNH_0413200 | 78 S | NPESSVGDN | 0.997 | unsp | PKNH_0413200 | 78 S | NPESSVGDN | 0.997 | unsp | PKNH_0413200 | 100 S | SSADSTSGT | 0.994 | unsp | PKNH_0413200 | 149 S | VPTSSSGDS | 0.995 | unsp | PKNH_0413200 | 153 S | SSGDSPVPA | 0.991 | unsp | PKNH_0413200 | 207 S | NSSNSEIEM | 0.993 | unsp | PKNH_0413200 | 507 S | TFDESEDDI | 0.992 | unsp | PKNH_0413200 | 838 S | TPCHSVEGQ | 0.995 | unsp | PKNH_0413200 | 855 S | GQEASREDT | 0.998 | unsp | PKNH_0413200 | 30 S | ESDSSSESH | 0.994 | unsp | PKNH_0413200 | 47 S | SAGPSQGDS | 0.996 | unsp |