

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PKNH_0413300 | OTHER | 0.685403 | 0.251273 | 0.063324 |
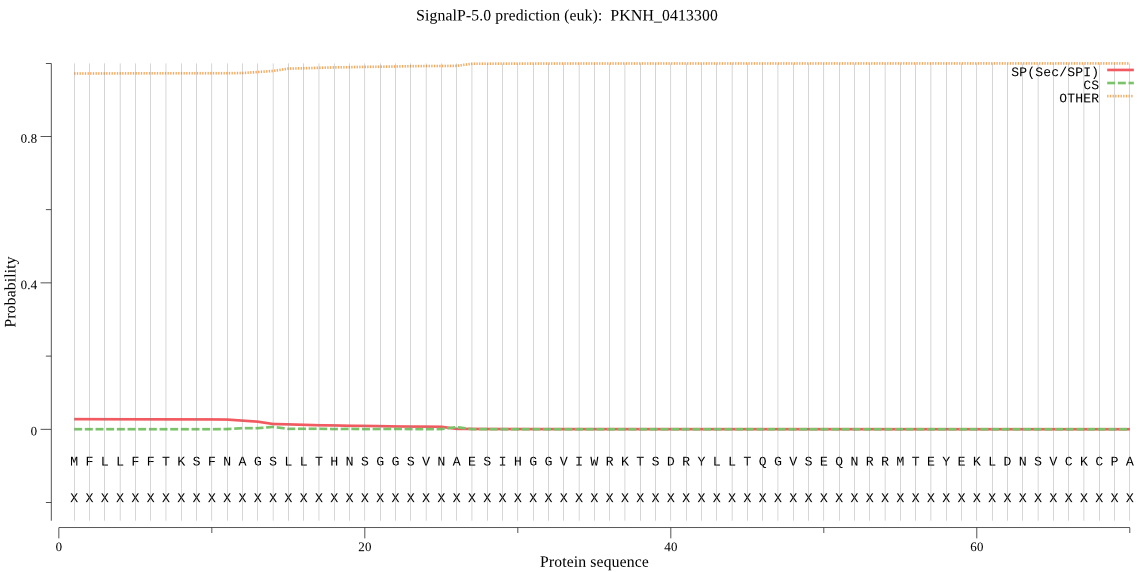
MFLLFFTKSFNAGSLLTHNSGGSVNAESIHGGVIWRKTSDRYLLTQGVSEQNRRMTEYEK LDNSVCKCPAANDPTSTIHGTSSKPGEPCEGLGQADNSKVKAHLLKKSIGLKINRMSGSE MEFLFIPHSYIYVTAQDSRARAFNILTYMEDNALPFTRKVYVPVETLTTTESSATETKWY RRALAENNDGPWDDEHDFFSKGMDSEEEEAIPPIGMDNVDPEEVQLRKTIDRILNWIYRI DEDGKKVLIMQEELDLSMKEDLINYCQIMKKVDKSGTLEEHQIGNEMHVFGNLIKLLQKH GDETVFTLRRKLRNPAMCMQNVKEWVLKRRGLVLPDSSSNDPSEHDKILDVTSKSSTSVR SNSRDASALRYSGEEEGMVDLEKLKMKCYHIFSLNGGVNTDGDHPQKSDGKDLCSPKDKE EVE
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PKNH_0413300 | 117 S | INRMSGSEM | 0.997 | unsp | PKNH_0413300 | 117 S | INRMSGSEM | 0.997 | unsp | PKNH_0413300 | 117 S | INRMSGSEM | 0.997 | unsp | PKNH_0413300 | 173 S | TTESSATET | 0.996 | unsp | PKNH_0413300 | 205 S | KGMDSEEEE | 0.997 | unsp | PKNH_0413300 | 257 S | ELDLSMKED | 0.996 | unsp | PKNH_0413300 | 343 S | SNDPSEHDK | 0.991 | unsp | PKNH_0413300 | 358 S | KSSTSVRSN | 0.996 | unsp | PKNH_0413300 | 372 S | ALRYSGEEE | 0.997 | unsp | PKNH_0413300 | 415 S | KDLCSPKDK | 0.998 | unsp | PKNH_0413300 | 56 T | NRRMTEYEK | 0.994 | unsp | PKNH_0413300 | 82 S | IHGTSSKPG | 0.995 | unsp |