

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PKNH_0413400 | SP | 0.002377 | 0.997622 | 0.000001 | CS pos: 16-17. TYG-NN. Pr: 0.3269 |
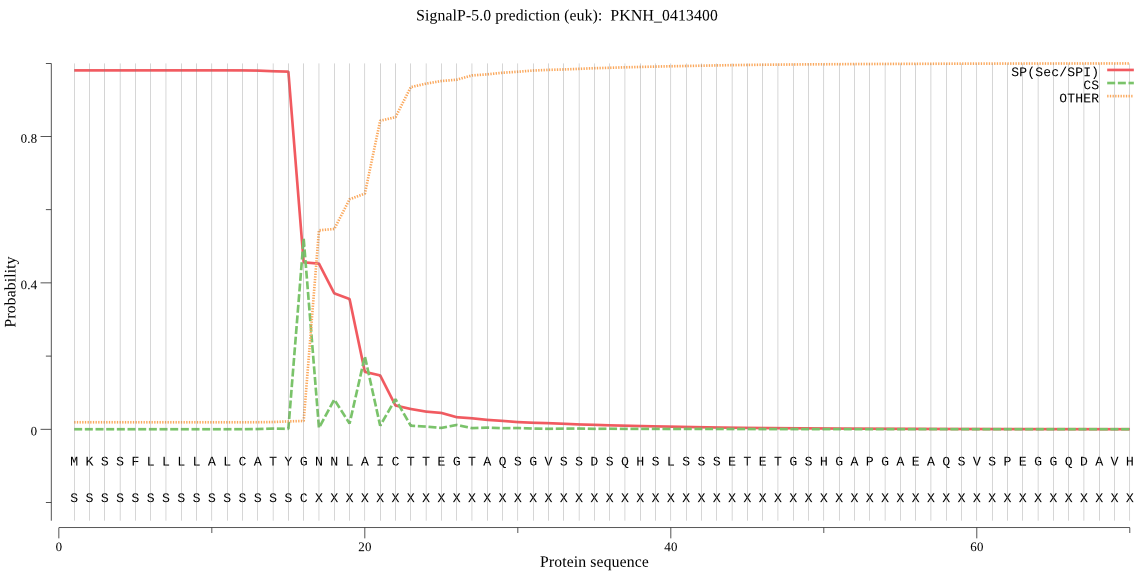
MKSSFLLLLALCATYGNNLAICTTEGTAQSGVSSDSQHSLSSSETETGSHGAPGAEAQSV SPEGGQDAVHSTNESAESDAESPTEPNPPQEDGTSNEDGNGGHSESSAPSVPGGSASPSS VENSNEQAAGTQLQVAPQKAQVKSALLKNFTGVKVTGPCDTEVGLFLIPYIYISVKVAED NIELSTNFPNSENIIVKFSNEGQEMNNKCRGDPKKTYKFIAYLDDNILTLKWVVYPPNGD GEKKVPDVKKYKLPNLERPITSIQVHSLMLQGENIIYTSKDYSIKNDIPETCEQVLSACF LSGNTDIENCYTCNLLLHKENTDDKCFDYVSADFKKEFEDIKLKGQDDEDSGEYKLAQAI NAILNGVYKTGANGKKELITSEELDANFMEQIGNYCQIMKKVDKSGTLEEHQMGTEVDVF NNLVKFLQKHQEEEYSTLEWKLQNPVLCLKNVNDWVMNKKGLLLPLLQNDDSHIHLGEYD KEGDEQKCCSSTYQEGDDGIIDLSVVHKNAHASSTPFTNHMFCNYDYCDRTKDTSSCLSK IEVQDQGNCATSWLFASKLHLETIKCMKGYKHIPSSALYVANCSNKETLNKDKCHTPSNP LEFLHTLEETKFLPSESDLPYCYKKVGNDICPEPKSHWKNLWTNIKLVDAQYQPNSISTK GYTEYQSKYFKGNMDAFIKLVKSEIINKGSVIAYVKAAGALSFDLNGKKVQNLCGEEGTP DLAVNIIGYGNYINEEGIKKSYWLLQNSWGYYWGNEGTFKVDMHGPDDCEHNFIHTAAVF NLDMPEFQSPIKDTELHSYYLKNSPDFYKNLYYNALDGECGNAPCNTVEGQDAPGEKATD QVGASGAGVATVTTTGTGAAPGTGAEAKAGAEAGAGAETEATKATEVLEPKEQQAQSQVT VVENSVSKDQPQPQPQPQPQPQPQLQQEQQQEQQQEQQQEQQQEQQQQQEQQQQQPQPPQ PALQDLTNEHSPHIGETEVKVEPEGENSNAELQKAKMVQIIHVLKHIKQTKMVTRVVTYQ GNYELGEHSCSRTEASSVEKLDECIKFCDEHYSECKLSISPGYCLTKKRNANDCNFCFV
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PKNH_0413400 | 82 S | SDAESPTEP | 0.995 | unsp | PKNH_0413400 | 82 S | SDAESPTEP | 0.995 | unsp | PKNH_0413400 | 82 S | SDAESPTEP | 0.995 | unsp | PKNH_0413400 | 95 S | EDGTSNEDG | 0.991 | unsp | PKNH_0413400 | 120 S | ASPSSVENS | 0.992 | unsp | PKNH_0413400 | 283 S | SKDYSIKND | 0.996 | unsp | PKNH_0413400 | 615 S | KFLPSESDL | 0.996 | unsp | PKNH_0413400 | 1037 S | TEASSVEKL | 0.997 | unsp | PKNH_0413400 | 39 S | DSQHSLSSS | 0.994 | unsp | PKNH_0413400 | 41 S | QHSLSSSET | 0.997 | unsp |