

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
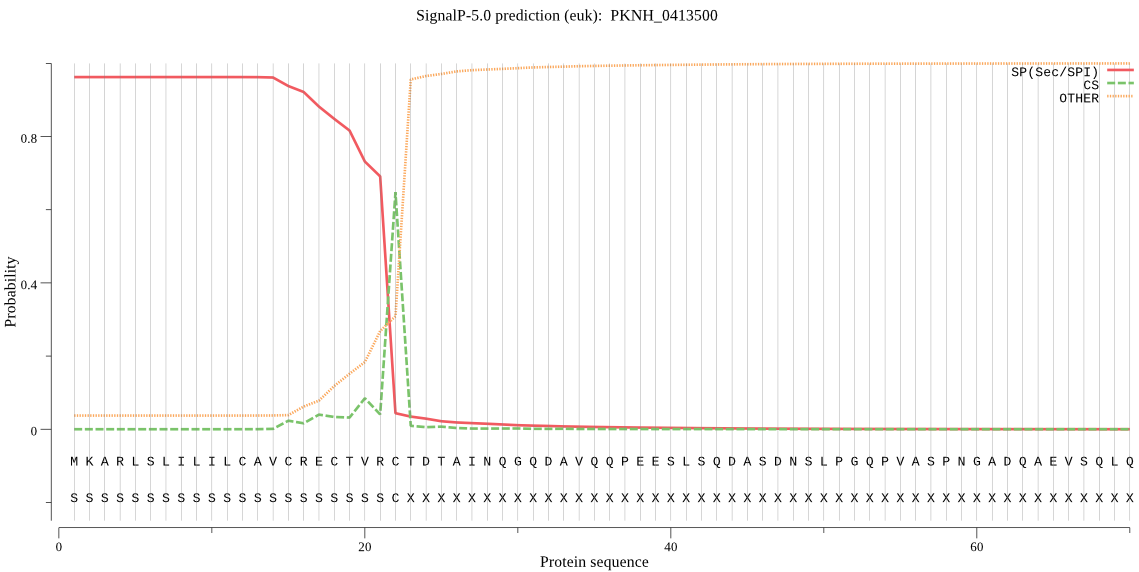
| PKNH_0413500 | SP | 0.016263 | 0.983634 | 0.000104 | CS pos: 22-23. VRC-TD. Pr: 0.8458 |
MKARLSLILILCAVCRECTVRCTDTAINQGQDAVQQPEESLSQDASDNSLPGQPVASPNG ADQAEVSQLQEGAVESSSNSADASNPNANADVTDVEGEKAATPSEGSKEGMQVKSSLLKG YKGVKVTGPCSASFLVFFAPYLFIDVDADSSNIYLGTDLNDLEITEKMGKGKDEKNKCQE GKTFKFVAFVVNDHLTIKWKVYDSEDQTPTPNSNVEMKKYKVKNLSGEFTSVQVHSTIQQ NGSNVFESKNYALSNDMPEKCDAIAANCFLSGSVYIEKCYRCTLKMEKVNSSDVCYNYIP KVDTPTKEEAPIYDHYLKNSPNLGGSTSNKNIADQEGASTINAAGGVHESVLQGQEAEEK EVKDVAVKAAPEGNPVVTEEKADGQVSTEVPSLPEAKVTEVQSAGTTGGEQPVPAVPEGQ AGAPSTTGPDSTSGTSPQPPVVSTQLSHVLKYIKKNKVKMNLITYKNNEAVSSGHDCSRS YSVNPDKYEECVKICEANWSKCENDAVPGFCLYQYAKENECFFCYV
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| PKNH_0413500 | 226 S | VKNLSGEFT | 0.993 | unsp | PKNH_0413500 | 431 S | TGPDSTSGT | 0.996 | unsp |