

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
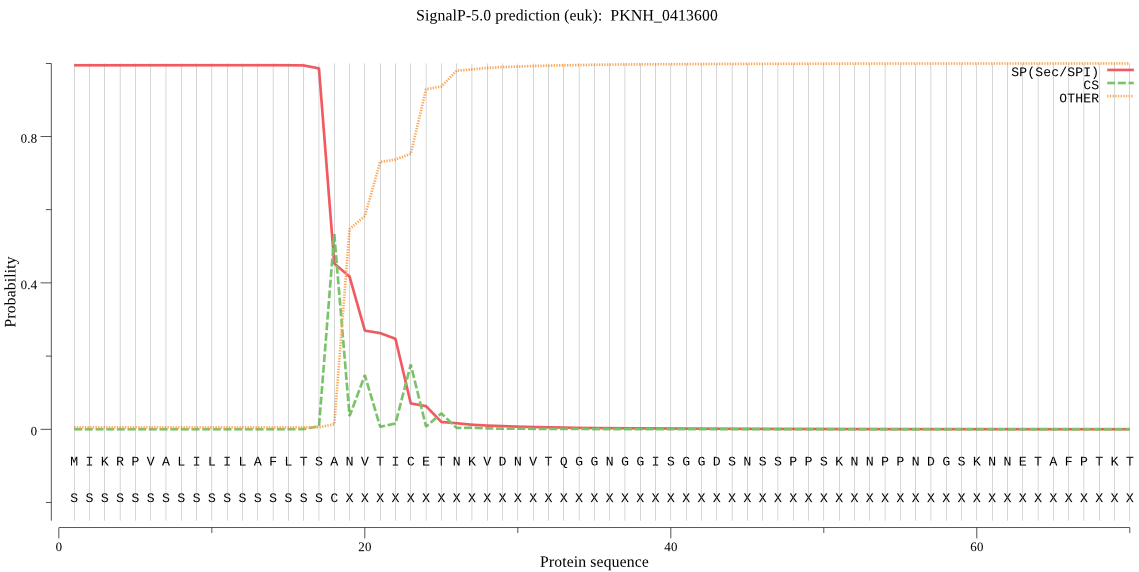
| PKNH_0413600 | SP | 0.000519 | 0.999480 | 0.000001 | CS pos: 23-24. TIC-ET. Pr: 0.6601 |
MIKRPVALILILAFLTSANVTICETNKVDNVTQGGNGGISGGDSNSSPPSKNNPPNDGSK NNETAFPTKTISFNMPSNLEGAGSKNDSAKSTDEAGARTDGSLPSSTNATEKSNTSSGKE AEHTDISSGEREKTEPSTNEVAGEGHQTAVAMFEGNKTKSNGSLDGSMQVESALLKNYKG IKVTGPCGSYFRVYLVPHILIYALTKNSIIQIESLFDDNTRIDFEYADNMVNKCEEGKNF KLILYLKDNILTLKWNVLPSLSADDSDSTTKADVRKYKLPSLERPFTSIQVYTADAKEGI IETKNYALGADIPDKCNAIATDCFLNGNINIDKCYQCTLLVQNSEQNSEECFNYVSKDLQ NQHNNAKVKGQDDLNPKEYELTESIDIILKNIYKVDKVKKKKVLIGMDDLDDVLKVELLN YCKLLKEMDVKGTLDNLDLGDEIDIFNNMIRLLSMHPKENIITLQDKLRNTAICLKNVDD WIENKRGLLLPDEDESSSLAKNPTQVDKEDESDEEEDHVKESELLNQDMYRKDKDGTIDL VKAGKELKLRSPYFKNSKYCNYEYCDRWKDKTSCISNIEVEEQGNCGLCWIFASKLHLET IRCMRGYGHFRSSALYVANCSRRNRKDICEVGSSPTEFLQIVRDTGFLPLESDHPYLHKN AGNECPKIKENWINLWGNSKLLSHKMYGHFMVYKGLISYQSRHFKDNMHIFIDLVKREVQ NKGSVIIYIKTKDVIGYDFNGNGVQSLCGDKTPDHGANIVGYGNYINVNGEKRSYWLIRN SWGYYWGNEGTFKVDMHGPPGCKYNFIHTAVVFKVNLGAVDIPNKDVDLNHTYFPKHNAD FFHSLYFNNYDDEADNQKKIFPKYRNMDYSGSGYKWKKVSKKWQISGQDEDITEGASSEE TSSNDSSVAPKTAKPSPSAEKKIQILHVLKHIDKAKIMRGLVKYENISETQNDHSCARAH SKNPEKIEECKLFCEENWNNCKNHYSPGYCLSKLYSGSNCYFCYV
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PKNH_0413600 | 117 S | SNTSSGKEA | 0.997 | unsp | PKNH_0413600 | 117 S | SNTSSGKEA | 0.997 | unsp | PKNH_0413600 | 117 S | SNTSSGKEA | 0.997 | unsp | PKNH_0413600 | 281 S | YKLPSLERP | 0.993 | unsp | PKNH_0413600 | 512 S | KEDESDEEE | 0.998 | unsp | PKNH_0413600 | 880 S | WKKVSKKWQ | 0.991 | unsp | PKNH_0413600 | 897 S | TEGASSEET | 0.997 | unsp | PKNH_0413600 | 898 S | EGASSEETS | 0.995 | unsp | PKNH_0413600 | 902 S | SEETSSNDS | 0.994 | unsp | PKNH_0413600 | 916 S | TAKPSPSAE | 0.993 | unsp | PKNH_0413600 | 88 S | SKNDSAKST | 0.997 | unsp | PKNH_0413600 | 91 S | DSAKSTDEA | 0.993 | unsp |