

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PKNH_0413700 | SP | 0.002709 | 0.997228 | 0.000063 | CS pos: 22-23. TRC-GG. Pr: 0.4997 |
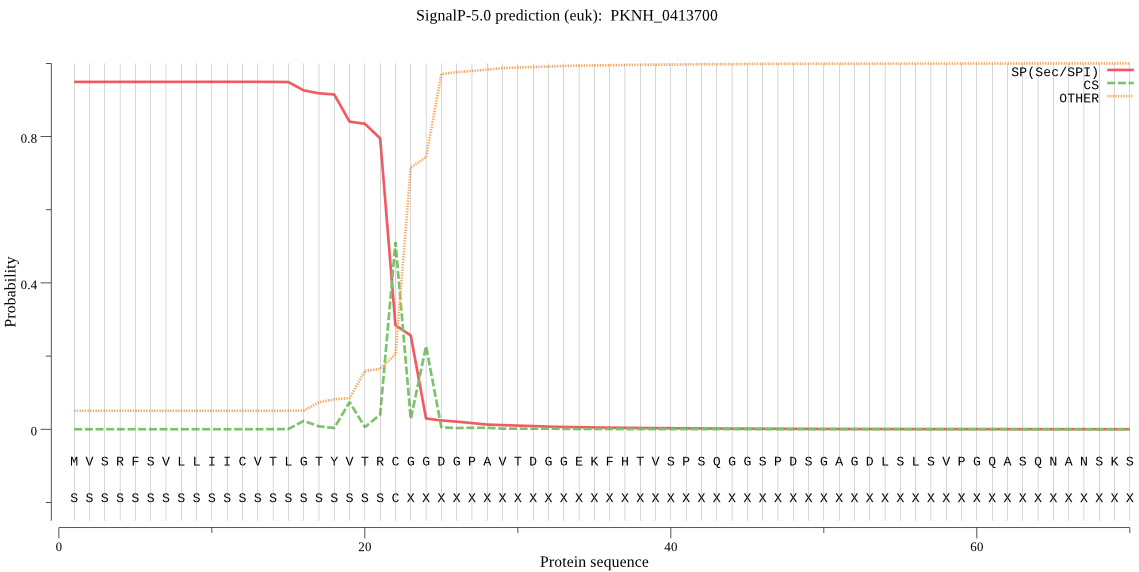
MVSRFSVLLIICVTLGTYVTRCGGDGPAVTDGGEKFHTVSPSQGGSPDSGAGDLSLSVPG QASQNANSKSNAMETASEGAVSMQVADPSEEDIQVTSALLKNYNGIKITGSCKAHFRVFL APHLWIYAAAPENKIQLRPNFGPITSIDLGNLINDCKKGANSNFKFVVHTRDNILTLKWK VTGKGESPGNEEDVKKYKLPSLERPFTSIQVHSANAKSKIIESKFYDLGSSMPAKCDLIA MNCFLKGSLDIEQCYHCTLLEKKMNEDSECFKYVSIEAKEFIKKDTPIKAQEEDANSAEH KLIESIEVILKAMYKSDKDYEKKELISLDQVDTNLKKELSNYCQLMKQVDTSGTLKNYKL ANEVETYRNLKKMLQMHKEENIITLQDKLRNAAICMKDIDKWIINKRGLTLPDGVTYSED NTKEYLSEELLKELEKEKSVFDDDVFDKDKNGVIDLNKVPSEMKFKSPYFKKSKYCNNEY CDRWKDKTSCISNIEVEEQGDCGLCWIFASKLHLETIRCMRGYGHFRSSALFVANCSNRD PGERCSVGSNPTEFLQIVKDTGFLPLESDLPYKYTDAKNSCPMNRNKWTNLWGNTKLLYH VRPNLFLKTLGYVSYESKFFRKNIDLFIDMLKREIQNKGSVIIYIKTEDNIDYDFNGKIV HSLCGHKDADHAANIVGYGNYINVNGEKSSYWLIRNSWGYYWGDEGNFKVDMYGPPGCKY NFIHTAVVFKIDLGIVEVPKKAEKPPHNYFAKYVPNFLYNLFFVCFDGGAADDDDIELGQ NGKVGAAGGEENAVVDGQAVVAGQTENTLSVASKNGAQPETNGSEVNAASVGKPNIASPM GSQMNAIAPQVGSVASTTNVAPLNVTVREGQHVGGAVTLTPVDNSGEKKIPVTHILKYVK ETKMTKTLVNYDSLSEIEDSHSCARTYAKDEVGQEECIQFCLKNWSSCKGHYNPGYCLTK MYTGNNCIFCSV
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PKNH_0413700 | 201 S | YKLPSLERP | 0.993 | unsp | PKNH_0413700 | 201 S | YKLPSLERP | 0.993 | unsp | PKNH_0413700 | 201 S | YKLPSLERP | 0.993 | unsp | PKNH_0413700 | 316 S | AMYKSDKDY | 0.997 | unsp | PKNH_0413700 | 427 S | KEYLSEELL | 0.992 | unsp | PKNH_0413700 | 439 S | EKEKSVFDD | 0.995 | unsp | PKNH_0413700 | 89 S | VADPSEEDI | 0.993 | unsp | PKNH_0413700 | 187 S | GKGESPGNE | 0.996 | unsp |