

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PKNH_0413800 | SP | 0.261645 | 0.735614 | 0.002741 | CS pos: 24-25. VKS-NI. Pr: 0.9778 |
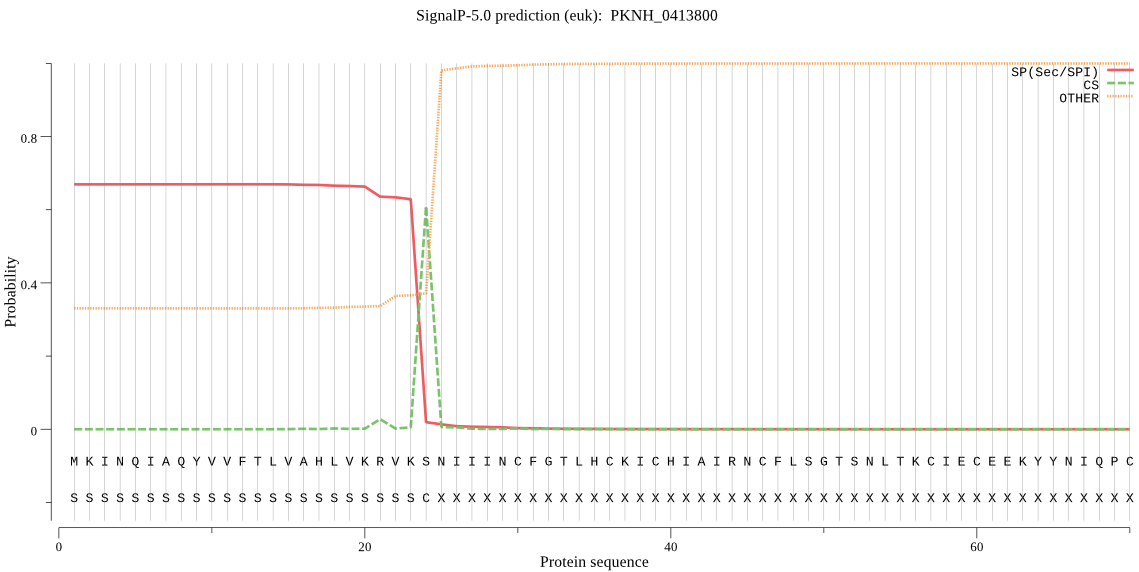
MKINQIAQYVVFTLVAHLVKRVKSNIIINCFGTLHCKICHIAIRNCFLSGTSNLTKCIEC EEKYYNIQPCTHHTENFLQISRDKGAFVEMKNHDYLTEAKVDDLISEIVKLSLERQKNAV PETEQTKQDFQKKIMQLCLYSNFSDHYENAKKHTQANAEEVEKHINKIVIMYMNESNNME HIINSLKNPALCLKDPAEWTKDRAGYKDNDVPSVGIIPERKLFKPYEVKSLMSSLYNYSS NCNRQFCDRFSDPNECENSIRVLNQGTCGNCWAFASSQVISAYRCRKGLGFAEPSIKYVT LCKNKHSNDLEENPFGHYNDNICREGGHLSYYLETLDRTKMLPTSYDVPYNEPIKGVDCP ESSDKWANMWEHVNPLTRILNGYLYRGYIKISFHEYAQSGKTDELISLIKDYIIEQGSLF VSMEVNEKLSFDHDGEKVMMNCEYRASPDHALTLIGYGDYLKPNGEKSSYWLIRNSWGSH WGDKGNFKIDMYGPSDCNGAVLCNAFPLFLQMNGDKITKPLPDDLASTDNKTRYDHSYFT RDRNRRASRRYNPNNERDQPNNQEDKDPFDNPNNNDNYDGNDRDWPPFDPFKDYVYPYMD NRKDAERDNGGISDNRSNKIRRTIFKTTLVVNIGDTNFKRNIYMKKKDEYKEQHSCLRTY SLDSQADVSCRNNCDQHIDLCKHYTSIGRCLMRFAPNYKCVYCGM
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PKNH_0413800 | 548 S | NRRASRRYN | 0.998 | unsp | PKNH_0413800 | 548 S | NRRASRRYN | 0.998 | unsp | PKNH_0413800 | 548 S | NRRASRRYN | 0.998 | unsp | PKNH_0413800 | 664 S | YSLDSQADV | 0.99 | unsp | PKNH_0413800 | 64 Y | CEEKYYNIQ | 0.99 | unsp | PKNH_0413800 | 112 S | IVKLSLERQ | 0.992 | unsp |