

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PKNH_0423400 | SP | 0.000523 | 0.999347 | 0.000130 | CS pos: 23-24. ICA-HK. Pr: 0.7466 |
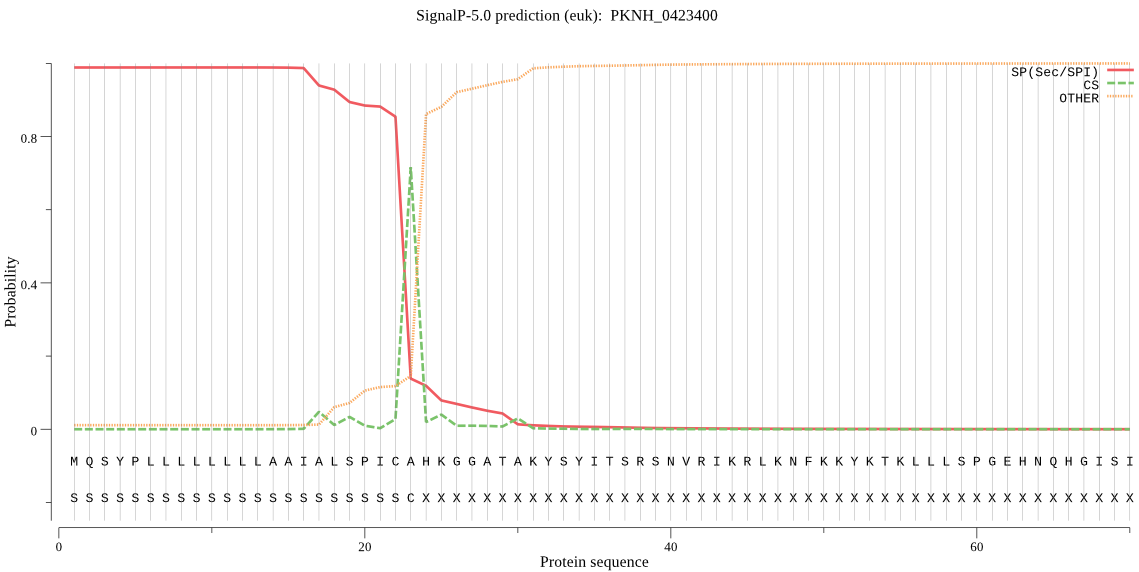
MQSYPLLLLLLLLAAIALSPICAHKGGATAKYSYITSRSNVRIKRLKNFKKYKTKLLLSP GEHNQHGISIPSMLLSKRIIFLSSPVYPHISEQIISQLLYLEYESKRKPIHLYINSTGDL ENNKIINLNGITDVISIIDVIEYISSDVYTYCLGKAYGISCILASSGKKGFRFSLKNSSF CLNQSYSVIPFNQATNIEIQNKEIMNTKRKVVEIIANNTGKDKSHIEQILERDRYFSAPE AVEFNLVDHILEKE
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PKNH_0423400 | 224 S | GKDKSHIEQ | 0.997 | unsp | PKNH_0423400 | 224 S | GKDKSHIEQ | 0.997 | unsp | PKNH_0423400 | 224 S | GKDKSHIEQ | 0.997 | unsp | PKNH_0423400 | 237 S | DRYFSAPEA | 0.996 | unsp | PKNH_0423400 | 59 S | KLLLSPGEH | 0.994 | unsp | PKNH_0423400 | 174 S | GFRFSLKNS | 0.996 | unsp |