

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PKNH_0501300 | SP | 0.421899 | 0.577032 | 0.001069 | CS pos: 23-24. LNG-YI. Pr: 0.8399 |
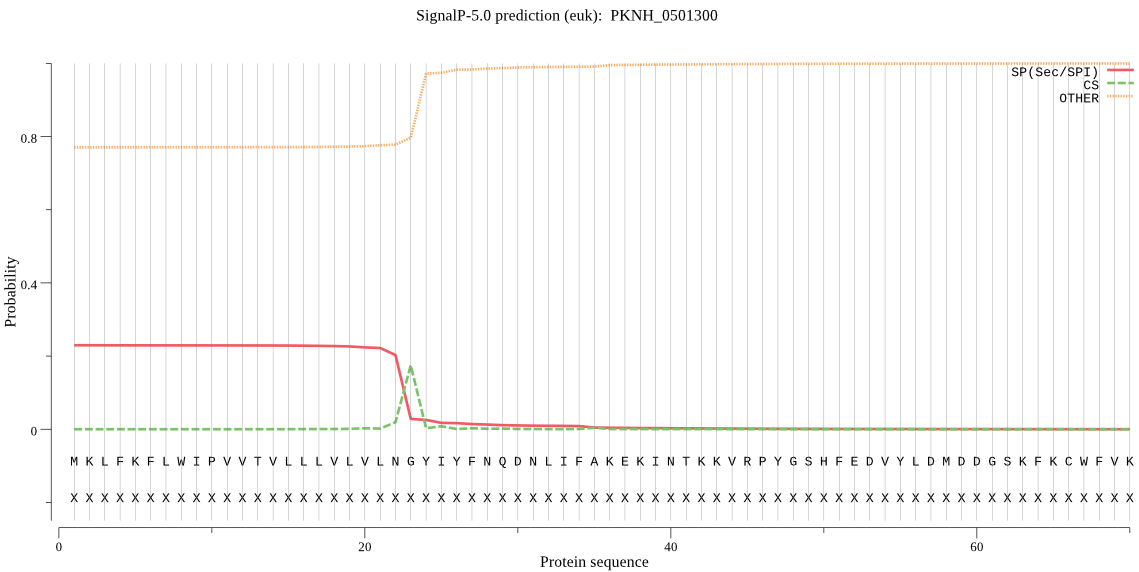
MKLFKFLWIPVVTVLLLVLVLNGYIYFNQDNLIFAKEKINTKKVRPYGSHFEDVYLDMDD GSKFKCWFVKTKDHEKKPVFLYYLGKGGYIEKYVKLFDMIVQRVDVSIFSCSNRGCGTNE GTPSEEQLYKDALVSFNYLKGKNTKQLFIFGNSMGGAVALETASRHQKDIYGVILENTFL SMKKISEETHPFLNFFLISFDTLIRTKMDNMEKIKKIHIPLLINISEQDEVVPPEHSKIL FELSPSKHKFKYLSKKGTHNNIIKVDDGSYHASLKKFVQTAISVREGKEIPEIERPPPAA AV
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PKNH_0501300 | 254 S | FKYLSKKGT | 0.993 | unsp | PKNH_0501300 | 254 S | FKYLSKKGT | 0.993 | unsp | PKNH_0501300 | 254 S | FKYLSKKGT | 0.993 | unsp | PKNH_0501300 | 283 S | QTAISVREG | 0.996 | unsp | PKNH_0501300 | 55 Y | FEDVYLDMD | 0.991 | unsp | PKNH_0501300 | 186 S | MKKISEETH | 0.991 | unsp |