

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
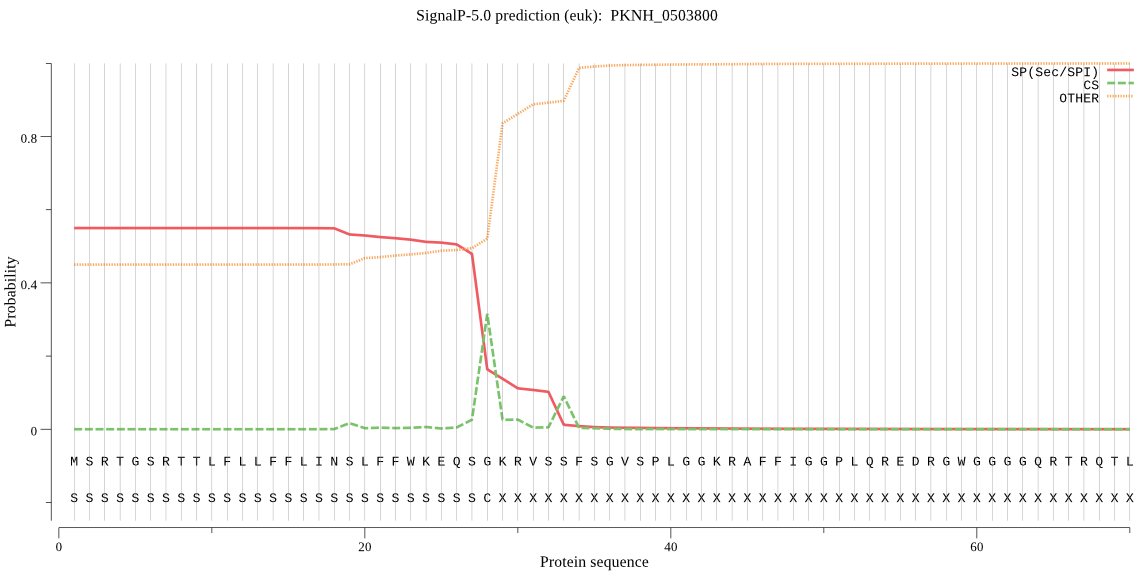
| PKNH_0503800 | SP | 0.072297 | 0.925803 | 0.001901 | CS pos: 28-29. QSG-KR. Pr: 0.7548 |
MSRTGSRTTLFLLFFLINSLFFWKEQSGKRVSSFSGVSPLGGKRAFFIGGPLQREDRGWG GGGQRTRQTLDGAQKTHIQADQALSTKRSAANGRLFYSLGRHPVEQISENGGACERLSVR QSPGLSARWNHPLRMSDEEYTINSDDYTEKAWEAITTLNKIGEKYESAYVEAEMLLLALL NDGPEGLAQRILKESGIDTDLLIQEIDEYLKKQPKMPSGFGEQKILGRTLQSVLGTSKRL KKEFHDEYISIEHLLLGIVAEDSKFTRPWLLKYNVNYEKVKKATERVRGKKKVTSKTPEL TYQALEKYSRDLTALARAGKLDPVIGRDTEIRRAIQILSRRTKNNPILLGDPGVGKTAIV EGLAIKIVQGDVPDSLKGRKLVSLDLSSLIAGAKYRGDFEERLKAILKEVQDAEGQVVMF IDEIHTVVGAGAVAEGALDAGNILKPMLARGELRCIGATTVSEYRQFIEKDKALERRFQQ ILVEQPSVDEAISILRGLKERYEVHHGVRILDSALIQAAVLSDRYISYRFLPDKAIDLID EAASNLKIQLSSKPIQLDNIEKQLVQLEMEKISILGDSPPRGGGSFSTVSSSSKDDDSTQ GGKDPPVDYSQSPNFLKKKINEKEINRLKMIDHIMSQLRKEQKSILESWSCEKTYVDNIR AIKERIDVVKIEIEKAERYFDLNRAAELRFETLPDLEKQLKKAEDNYLSDIPEKNRMLKD EVTSEDIMNIISLSTGIRLNKLQKSEKEKILNLENELHKQIIGQDDAVRIVSKAVQRSRV GMNDPKRPIASLMFLGPTGVGKTELSKVLADVLFDTPDAVIHFDMSEYMEKHSISKLIGA APGYVGYEQGGLLTDAVRKKPYSIILFDEIEKAHPDVYNLLLRVIEEGKLTDTKGNLANF RNTIIIFTSNLGSQSILDLANDPNKKDKIKEQVMKSVRETFRPEFYNRIDDHVIFDSLSK KELKQIANIEIEKVANRLIDKNFKISIDDAVFSYIVDKAYDPAFGARPLKRVIQSEIETE IAVRILNETFVENDTIRVSLKDGGLHFSKG
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PKNH_0503800 | 136 S | PLRMSDEEY | 0.996 | unsp | PKNH_0503800 | 136 S | PLRMSDEEY | 0.996 | unsp | PKNH_0503800 | 136 S | PLRMSDEEY | 0.996 | unsp | PKNH_0503800 | 195 S | ILKESGIDT | 0.997 | unsp | PKNH_0503800 | 375 S | DVPDSLKGR | 0.991 | unsp | PKNH_0503800 | 527 S | DRYISYRFL | 0.993 | unsp | PKNH_0503800 | 592 S | TVSSSSKDD | 0.998 | unsp | PKNH_0503800 | 593 S | VSSSSKDDD | 0.998 | unsp | PKNH_0503800 | 745 S | KLQKSEKEK | 0.996 | unsp | PKNH_0503800 | 936 S | QVMKSVRET | 0.997 | unsp | PKNH_0503800 | 959 S | FDSLSKKEL | 0.997 | unsp | PKNH_0503800 | 986 S | NFKISIDDA | 0.993 | unsp | PKNH_0503800 | 1015 S | RVIQSEIET | 0.991 | unsp | PKNH_0503800 | 1039 S | TIRVSLKDG | 0.998 | unsp | PKNH_0503800 | 85 S | DQALSTKRS | 0.995 | unsp | PKNH_0503800 | 118 S | CERLSVRQS | 0.995 | unsp |