

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PKNH_0602500 | SP | 0.067632 | 0.931917 | 0.000451 | CS pos: 28-29. GGS-ER. Pr: 0.6689 |
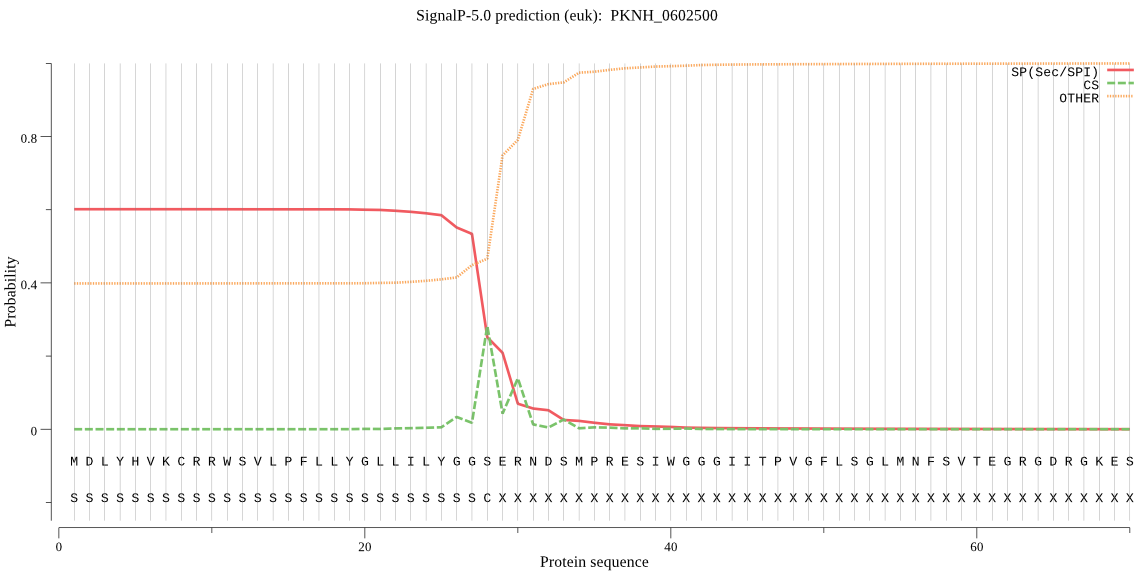
MDLYHVKCRRWSVLPFLLYGLLILYGGSERNDSMPRESIWGGGIITPVGFLSGLMNFSVT EGRGDRGKESTFAKEDHKINNRISRRVISSNGEGFHATVYFSGRNNGREELHSEDESVNR WDGIPDKVSQFMHKMINVKYENLMEDARRGGLLLERNEISHFGWKANEAINLSPMDEEES IKWKWKKEKEESASVKFGPNNKCFFLSRKEYSRMGRILSSANEDHPEESTIGVPNGDDTF EGSVLNIHFTENRNLYTNVKVGGQPLKLALNSRVEGIYVFMKDSQACYVGEEGMNRICYD PKTSRNSTWCNNDLLCLPAILSRPYECYSDSNLLLENKAEYPSMYYDSLKFTESHVEGSD DVELLDWRRDDRATNETSSANPINNLVRRGEDNTFQNTDIKLITDLSVYNGWSLFKDTDG MMGLAGRELSCRHVSAWNSIIEKNKSLYALDVNLPEGSVKPFVESASQNDSKKVGSSYGK FVSKKASTKKGDAATERNGTISEIHIGDYKKNFGPIVWSEARERGGIFSDSFMQFTLYNL QVCENNIFGKYSSNWQGVIDLSSKCLVLPKMFWLSLMEYLPVNKNDERCIPKNKEVNFDE STIPRMCSVDPRSRPLPVLKFYLSDNDIVSDNNVGGMNGASEEASQRRGIEQVHIPLDNL IINEEGQNDGYLCVLPDVHEGVSSENSGRTTKPLIKFGTYVINNLYVVVDQENYKVGFVN KKDYHFTNDRCTQRPVCIGNQFYEPALNICVDPDCSVWYFYTLNEETKKCESISSRFYVF LFILLLLLFMDIQSYYFYRKSVHVAKVSSR
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PKNH_0602500 | 173 S | AINLSPMDE | 0.993 | unsp | PKNH_0602500 | 173 S | AINLSPMDE | 0.993 | unsp | PKNH_0602500 | 173 S | AINLSPMDE | 0.993 | unsp | PKNH_0602500 | 194 S | EESASVKFG | 0.996 | unsp | PKNH_0602500 | 467 S | VESASQNDS | 0.995 | unsp | PKNH_0602500 | 487 S | SKKASTKKG | 0.996 | unsp | PKNH_0602500 | 33 S | ERNDSMPRE | 0.993 | unsp | PKNH_0602500 | 113 S | EELHSEDES | 0.997 | unsp |