-
-
-
Computed_GO_Function_IDs: GO:0008233
-
-
Computed_GO_Process_IDs: GO:0006465
-
-
Curated_GO_Component_IDs:
-
Curated_GO_Components:
-
Curated_GO_Function_IDs:
-
Curated_GO_Functions:
-
Curated_GO_Processes:
No Results
-
Fasta :-
>PKNH_0702100
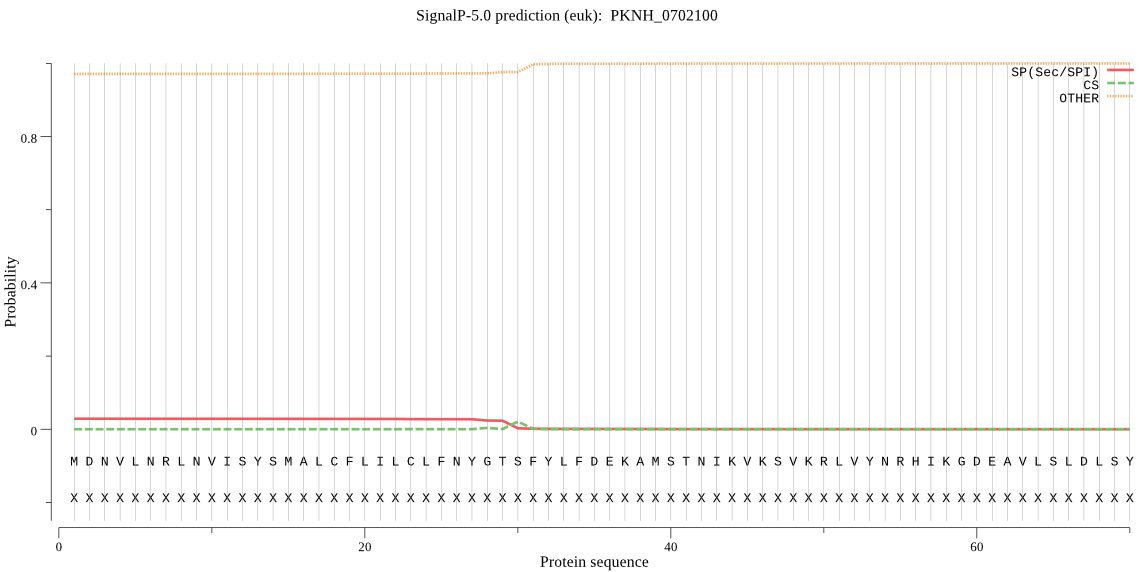
MDNVLNRLNVISYSMALCFLILCLFNYGTSFYLFDEKAMSTNIKVKSVKRLVYNRHIKGD
EAVLSLDLSYDMSKAFNWNLKQLFLYVLVTYETPEKVKNEVIIQDYIITNKNVAKRTYKN
FITKYSLKDYNNGLRNNNINLQVCYKYMPIVGLSRSYEGTKISYKLPAEYFDNLPSNYPL
YYPDK
- Download Fasta
-
MitoProt II - v1.101
File : /home/rajan/sadaf/4480_mitoprot/test/PKNH_0702100.fa
Sequence name : PKNH_0702100
Sequence length : 185
VALUES OF COMPUTED PARAMETERS
Coef20 : 4.546
CoefTot : -0.029
ChDiff : 8
ZoneTo : 34
KR : 1
DE : 1
CleavSite : 0
HYDROPHOBIC SCALE USED
GES KD GVH1 ECS
H17 : 1.888 2.194 0.448 0.686
MesoH : -0.777 0.360 -0.431 0.171
MuHd_075 : 26.939 13.656 5.933 5.073
MuHd_095 : 23.712 15.726 5.505 4.978
MuHd_100 : 26.401 19.679 7.909 5.077
MuHd_105 : 27.177 23.197 9.097 5.012
Hmax_075 : 12.600 15.983 2.191 5.437
Hmax_095 : 17.800 18.200 3.026 6.107
Hmax_100 : 19.000 20.200 4.565 6.360
Hmax_105 : 4.900 13.900 1.974 2.310
CLASS NOT-MITO MITO(/CHLORO)
DFM : 0.8962 0.1038
DFMC : 0.8521 0.1479
-
Fasta :-
>PKNH_0702100
ATGGACAACGTGCTGAATAGGCTAAATGTCATTTCGTATTCCATGGCCTTGTGCTTTCTT
ATCCTATGCCTCTTCAACTATGGAACTTCCTTTTATTTATTTGACGAGAAAGCAATGTCG
ACGAATATAAAAGTAAAGAGTGTGAAGAGGTTGGTGTATAATCGACACATAAAAGGAGAC
GAAGCCGTGCTATCGCTAGACCTATCATATGACATGAGCAAAGCATTCAATTGGAATTTG
AAGCAGCTATTTCTTTATGTTCTAGTAACGTATGAAACACCAGAAAAGGTAAAAAACGAA
GTTATTATACAGGATTATATAATTACCAATAAAAATGTGGCCAAAAGAACCTACAAAAAT
TTCATCACGAAATATTCTCTTAAAGATTATAATAATGGATTAAGAAATAATAATATTAAT
TTGCAAGTTTGTTACAAATATATGCCTATAGTTGGTTTGTCAAGATCTTATGAGGGTACT
AAAATTTCGTACAAATTGCCTGCAGAATATTTTGACAATTTGCCGTCCAATTATCCTTTG
TATTACCCAGACAAGTGA
- Download Fasta
|
ID | Site | Peptide | Score | Method |
|
PKNH_0702100 | 47 S | IKVKSVKRL | 0.992 | unsp |


