

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PKNH_0711400 | SP | 0.209362 | 0.788978 | 0.001660 | CS pos: 18-19. VGC-FN. Pr: 0.6932 |
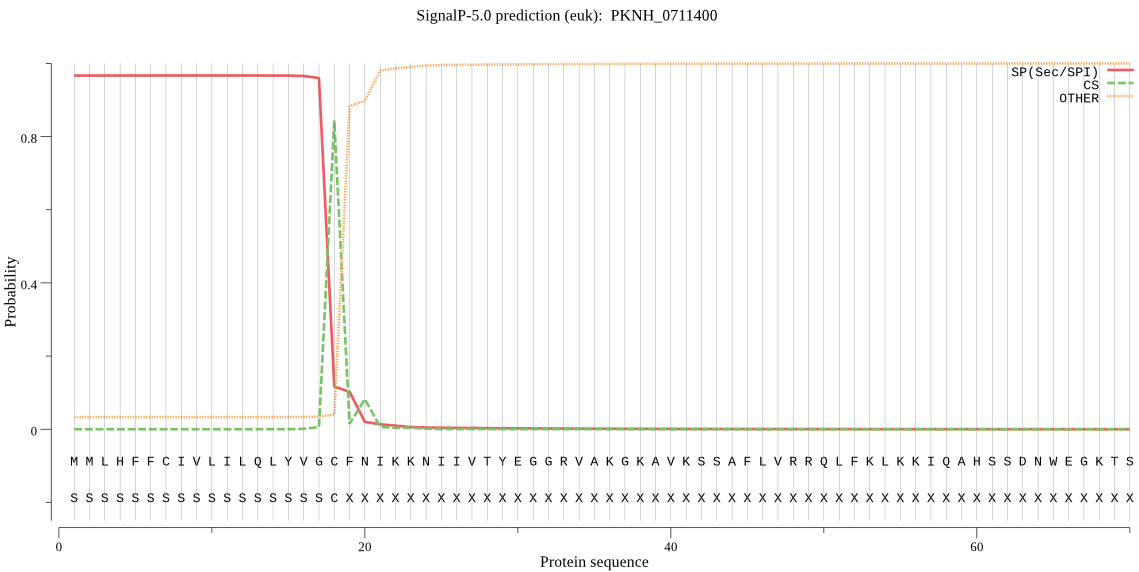
MMLHFFCIVLILQLYVGCFNIKKNIIVTYEGGRVAKGKAVKSSAFLVRRQLFKLKKIQAH SSDNWEGKTSIDEAQNNEAKNTLREFYDRNFRRLRKTFEFVKYGLISIQNNLLSNSLASQ LAISVSFFFLHFYMLSRHFLILFPYQLIPNHSNILMSLDMNNTLFFLLSIYSFKRFKDHF HSFKQRLQLNRFTIRLQDNKRKNILSICFFLIASYVLSGYVSIYTERILTLWKLFSRPLS ENVVKSLQILTGHFIWVGCSIVVFRKLLYPYFLNNESNLNLRHKSSWWFEVIYGYVFSHF VFNIVDLLNNFIINRFMGEGEDEVYVDNSIDDIVSGREFVSTLLCIISPCFSAPFFEEFI YRFFVLKSLNLFMHIHYAVTFSSLFFAIHHLNIFNLLPLFFLSFLWSYIYIYTDNILVTM LIHSFWNIYVFLSSLYS
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| PKNH_0711400 | 325 Y | EDEVYVDNS | 0.992 | unsp | PKNH_0711400 | 335 S | DDIVSGREF | 0.991 | unsp |