

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
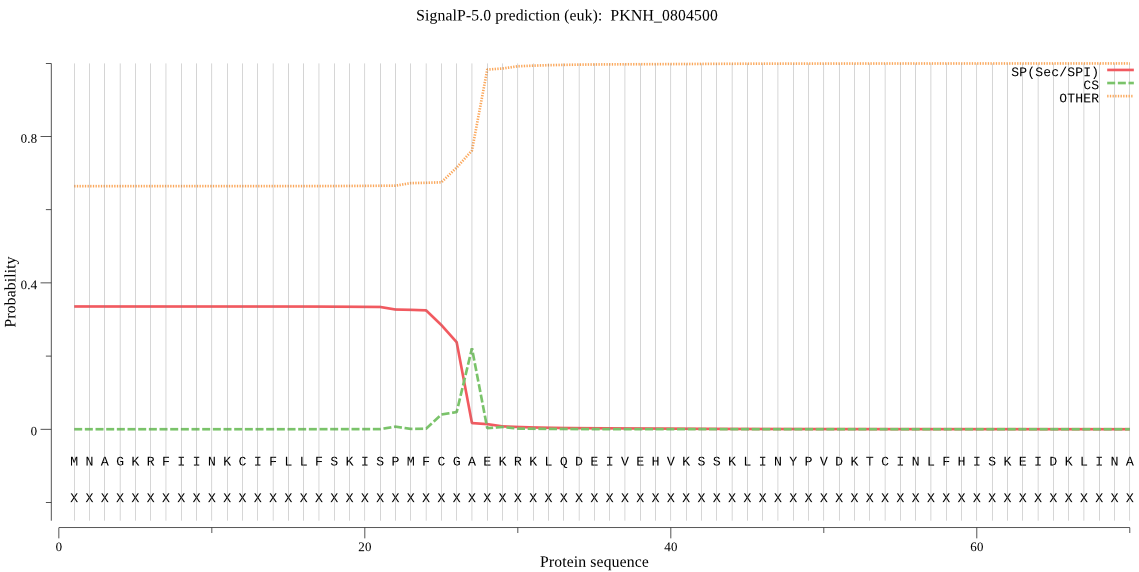
| PKNH_0804500 | SP | 0.418671 | 0.567251 | 0.014077 | CS pos: 25-26. MFC-GA. Pr: 0.4944 |
MNAGKRFIINKCIFLLFSKISPMFCGAEKRKLQDEIVEHVKSSKLINYPVDKTCINLFHI SKEIDKLINARSVFLSRNDQNGSHSSLQNCLNLYYFLITCAKLKKITFFHRKVLEYVEDR IATSLQIAGVDRAHEKYIPYSIHIKNEKSVSHLVAELHTDCAAYLKKINSVKIRYLTIDR NQINIDEEWKQKHQCAFNKKKLKIPISKHNYEFILNAVTEPALRQKVLTLLNSPYEKKRS LNKEVIKILKKRYDVATKLGYRNWVDYSMSHFTSEKGKYEKLRDFFNVIKRKVDRDYDRV SVHMGKLLSSSGVGGNQPGDRYGAHDDRRELRRDRLSPIRDKPSMQEWSYYYNKSMNKSD EYFVNTFFPHGYVWKNFMEIVSRIYNFSYEEVQEKNVTKKWPKGSLVYKIERRGSVNHAS AANSQANGKLVRSPVGQNPRREVVSPGDTLLGYMYIVPYMEIKIRDYFRVPSMNCLSNTC LIFTGHVVLDYKFIGTLPIEKKLFSCSEILTLFHEFGHAYHLLLLSRKYGLYQFSNMPLD YAEFFSHMNEHLVNNFSVLQALSNKADNNSKINEDIFKSIKFDSSRLANIYTHAVIDYEI HNLNPHDFFTRHLDNPGEVPYDHSYDHACGDNSLVPFYNLLENYLPYKIGSSYSIHSTSF PYHFSCYYAGSVLSYLLAEMRVLMNMSTGDLSMKRNSSLVSFQKKFYGIVAQDFVGSSGW SGSSVIDRVLANRDLLA
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PKNH_0804500 | 415 S | ERRGSVNHA | 0.996 | unsp | PKNH_0804500 | 415 S | ERRGSVNHA | 0.996 | unsp | PKNH_0804500 | 415 S | ERRGSVNHA | 0.996 | unsp | PKNH_0804500 | 445 S | REVVSPGDT | 0.998 | unsp | PKNH_0804500 | 692 S | TGDLSMKRN | 0.996 | unsp | PKNH_0804500 | 301 S | YDRVSVHMG | 0.996 | unsp | PKNH_0804500 | 337 S | RDRLSPIRD | 0.998 | unsp |