

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
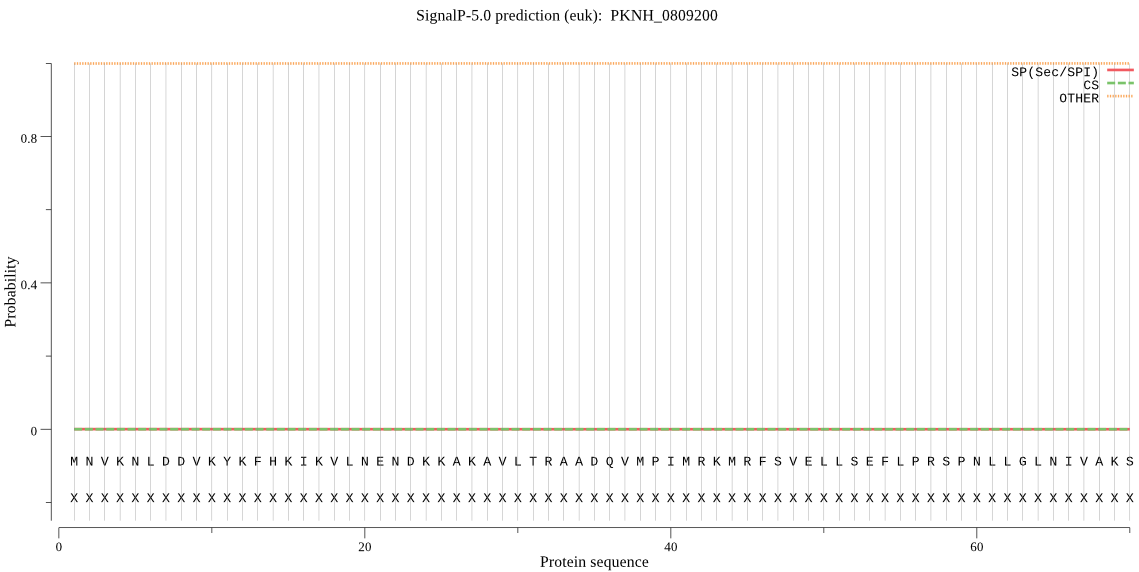
| PKNH_0809200 | OTHER | 0.999926 | 0.000024 | 0.000050 |
MNVKNLDDVKYKFHKIKVLNENDKKAKAVLTRAADQVMPIMRKMRFSVELLSEFLPRSPN LLGLNIVAKSEIKIRIRKKRGGELFHFNDIMGTLLHELAHIVHSGHDRSFYELLDKLVLE YNKLYTFGKAGNQINGGKKTGGSDFRICNGSPKFMAAQAAEMRLLNNFMNKDGEILNMSL ESCLTPEQYNNLFKNRKERDDKICSISNDTIVIDCLMDSTNHDDGENAKTSQNTKNDFKS SNSLQTRNKNVFISSEDCIKEEKQTHVPEQDVYKKSLNIPGAKRNKRQAAFEQWSGDKVI TLPDNIGATPKNNGSDCVHIVKVKRDCSGMKHPENENDSTVSDNVGIRKGKKRKVIILD
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| PKNH_0809200 | 47 S | KMRFSVELL | 0.993 | unsp |