-
-
-
Computed_GO_Function_IDs: GO:0008233
-
-
Computed_GO_Process_IDs: GO:0006465
-
-
-
-
Curated_GO_Function_IDs:
-
Curated_GO_Functions:
-
No Results
-
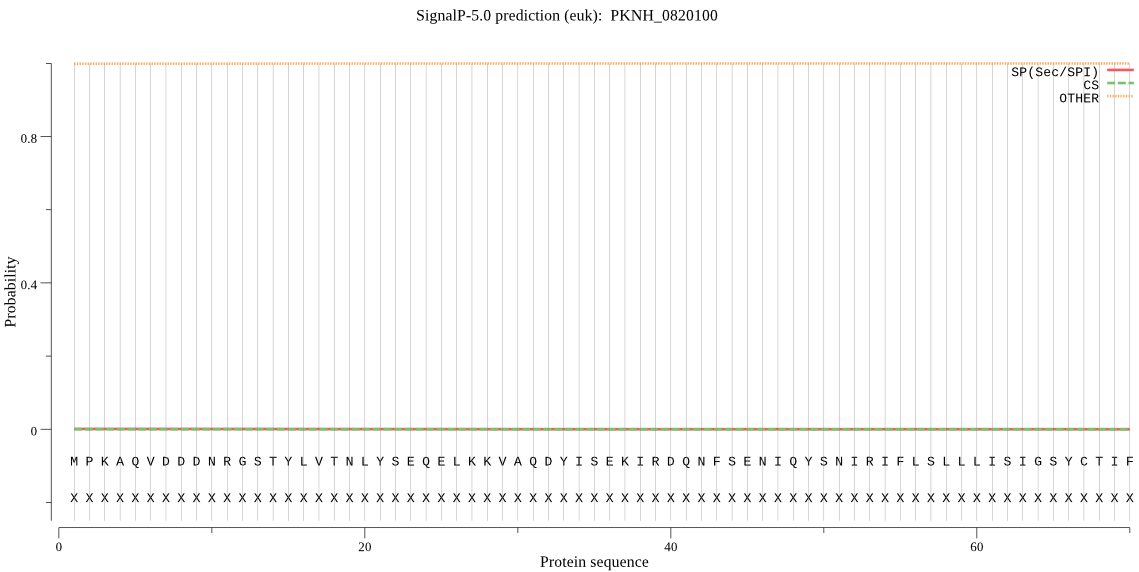
Fasta :-
>PKNH_0820100
MPKAQVDDDNRGSTYLVTNLYSEQELKKVAQDYISEKIRDQNFSENIQYSNIRIFLSLLL
ISIGSYCTIFVQYKKEPLLMIKLLIAFFIVSTILFFWEIVFFEDIFMILRTNNGGVVKLF
FELDIQKSALVLTYKMNKQKHSTSFELRKLFNEDGFLVQNYADAMLKQFISNHGKIFKLC
DNKKKA
- Download Fasta
-
MitoProt II - v1.101
File : /home/rajan/sadaf/4480_mitoprot/test/PKNH_0820100.fa
Sequence name : PKNH_0820100
Sequence length : 186
VALUES OF COMPUTED PARAMETERS
Coef20 : 3.433
CoefTot : -0.380
ChDiff : 4
ZoneTo : 6
KR : 1
DE : 0
CleavSite : 0
HYDROPHOBIC SCALE USED
GES KD GVH1 ECS
H17 : 2.153 2.653 0.492 0.904
MesoH : 0.491 1.332 -0.093 0.510
MuHd_075 : 8.391 8.150 1.751 1.687
MuHd_095 : 23.170 12.486 5.167 5.002
MuHd_100 : 17.458 9.781 4.100 4.274
MuHd_105 : 10.006 7.977 2.515 2.956
Hmax_075 : -2.800 3.587 -2.749 0.998
Hmax_095 : -1.312 4.112 -2.175 1.864
Hmax_100 : -4.900 2.800 -2.622 1.560
Hmax_105 : -2.800 4.667 -1.597 2.007
CLASS NOT-MITO MITO(/CHLORO)
DFM : 0.9482 0.0518
DFMC : 0.9725 0.0275
-
Fasta :-
>PKNH_0820100
ATGCCGAAAGCCCAAGTGGACGATGATAATAGGGGTAGCACATACCTCGTCACCAACCTG
TACAGCGAACAAGAGCTGAAAAAAGTTGCGCAGGACTACATAAGCGAAAAAATAAGGGAC
CAAAATTTTTCAGAGAATATCCAGTACTCTAACATTCGAATTTTTCTAAGTTTGCTTCTC
ATATCGATTGGATCCTACTGCACCATCTTCGTGCAGTACAAAAAGGAACCGTTGCTTATG
ATAAAATTGCTCATCGCCTTCTTCATCGTCTCGACAATTCTCTTCTTTTGGGAGATTGTC
TTTTTCGAAGACATCTTCATGATTTTGAGGACCAACAATGGCGGAGTTGTGAAGCTGTTC
TTCGAGCTGGACATCCAGAAGAGCGCCCTCGTATTAACCTACAAGATGAATAAACAAAAG
CATAGCACATCGTTTGAGCTTCGCAAGCTGTTCAACGAAGATGGATTCCTCGTCCAGAAT
TACGCAGACGCCATGTTGAAGCAGTTCATTTCAAATCACGGGAAGATTTTCAAACTCTGT
GATAATAAGAAAAAGGCCTGA
- Download Fasta


