

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PKNH_0827800 | SP | 0.211669 | 0.631660 | 0.156671 | CS pos: 23-24. EHA-RV. Pr: 0.2999 |
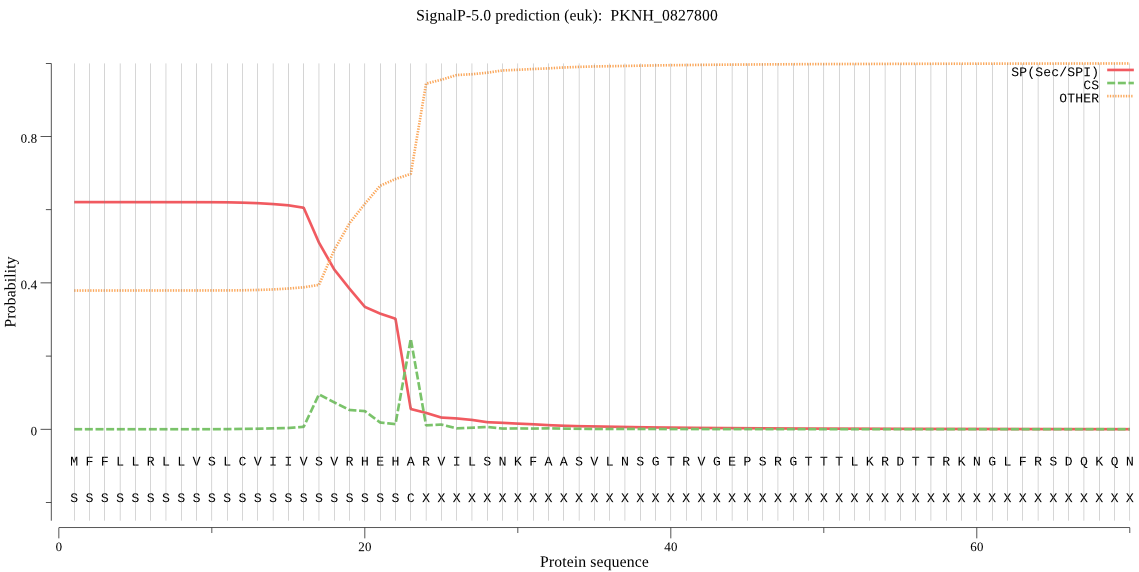
MFFLLRLLVSLCVIIVSVRHEHARVILSNKFAASVLNSGTRVGEPSRGTTTLKRDTTRKN GLFRSDQKQNTTSEHGYYYFTKEKNKIIKKKKKKFSLNYSRNPKVVTRGTRKKTHYPLLF ISPSSTSLNEITESVKALFNLDYIENSYLYSYIKSFKRTPPLTKLYLLCTFIFSIFMHAN KNIYKLIVFDFGKIFKEGQVWRLITPYFYIGNLYLQYFLMFNYLHIYMSSVEIAHYKNPE DLLTFFSFGYLSNLLFTIIGSMYNENVLNLQGYVNKLRRLILLGGKKSSISTKANTADVI ISKQQYNHLGYVFSTYILYYWSRINEGTLINCFELFLIKAEYVPFFFIVQNILLYNEFSL FEVASILSSYFFFTYEKNLQLNFLRHFNLALLKVLGIYPMYEAYKEEYE
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| PKNH_0827800 | 110 T | VTRGTRKKT | 0.99 | unsp | PKNH_0827800 | 291 S | KSSISTKAN | 0.996 | unsp |