-
Computed_GO_Component_IDs:
-
Computed_GO_Components:
-
Computed_GO_Function_IDs:
-
Computed_GO_Functions:
-
Computed_GO_Process_IDs:
-
Computed_GO_Processes:
-
Curated_GO_Component_IDs:
-
Curated_GO_Components:
-
Curated_GO_Function_IDs:
-
Curated_GO_Functions:
-
Curated_GO_Processes:
No Results
-
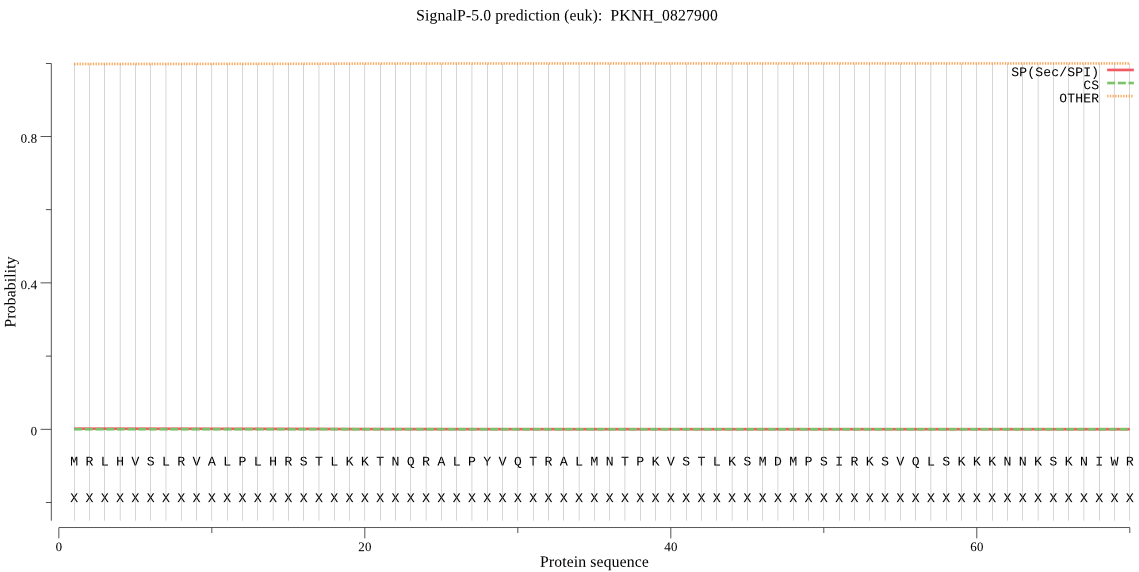
Fasta :-
>PKNH_0827900
MRLHVSLRVALPLHRSTLKKTNQRALPYVQTRALMNTPKVSTLKSMDMPSIRKSVQLSKK
KNNKSKNIWRDYLYVVIFKLYNNQVGNYVDLMIILKLLSKQISLEKKVLNFICEKLENHM
YRLTIRELSLLMLVLRKHKFDNLYFLNLLAKSILIKMNKNLSYKDLALITFSLSRSISLT
EKVYTNEIFHLSVVKIHNDLKNLNFHSLTLFFYSYSLYFLHHFEHFNSFFYMTKNFIQMI
KKNMYLFNPTDLMFTFLSAIHIYDSFNNGVRNVQCDSVRISSGAEVSLTYATLPRGEPTN
NYALTNYTETYDQPTLVPSKMTRAENPATMSATTEALHQANYPNEKVEKQDGKAREETLS
NFLTQVRYAIVEKLKCFKVEEVTNVLFASIDRDFTINKRYFPFLQKDKIHIEDYLHVYVK
KGQMWEEQHVERGELPHCRANPPAEESSHVPPQPTNYIIRISQGNHQEESFIHALSEEII
YRNEFLTARQILLLLYALRGNNTPFIQLEKILLKRLVCLEKELTVSQVMFVYQYLYFRTC
LFHELRRYAPKVYKHDRGKKEVFLLCDGDSATGEISQQEDQSTLPSERKNEKDNKVYYLH
DDTSHLKKRRKRDGTKIFLYDNFIFKYPAQVVSTEGEEVANLQEKKELKNFISMKRRRDS
LSEVNRSTVRNDEDKILTGETEYQITCYNFYLDVYKIVCLKMLNFLKNENINTCTILHVL
DIFKKLNIRDYRMIRHLYKMEKDILFLDNANLEKVLKILLNFNLYNIMHYLGVHRILQFI
NFNSVDECVSILGILGMMSTRKKTGHHEGREKAFPNFPTACTENAINYILKNLKDLSNIN
ALEKIQITPVYNSLPLKYFKLFKNVRSVRGLAQLWFQQNGGKKNHLAWSGENDPNRIRNF
VLHFVDDVNLGKVPFGSNGSCRLDYGGANQMRQTPNATAQVNCITEYRDIEEEIFQDLHT
IFPQTVEVYKNVNVSNYTFPMAINLLTLGGAIPNRSSQANATDLVKKHSLGNQCGSDRPM
RIQELDRKNTLLVDILYGQDFYYCLSGNKEQQLKKHNIYVAYYGMHANKTNKKMVNYKKY
SILKCIRKSGYHFICIDAKRYVKNKKKNRDNHLNRDYLTSLILDLIKRKKRIPLGINRRV
SRKNTSGTSSRKRRRRRPLLREKRHTCSVQGVISHVETNKYLHKVHTGGEKVSFPGTNHL
EKLRYMFQLG
- Download Fasta
-
MitoProt II - v1.101
File : /home/rajan/sadaf/4480_mitoprot/test/PKNH_0827900.fa
Sequence name : PKNH_0827900
Sequence length : 1210
VALUES OF COMPUTED PARAMETERS
Coef20 : 4.618
CoefTot : -2.115
ChDiff : 66
ZoneTo : 104
KR : 20
DE : 3
CleavSite : 54
HYDROPHOBIC SCALE USED
GES KD GVH1 ECS
H17 : 1.376 1.271 0.108 0.574
MesoH : -0.067 0.481 -0.429 0.285
MuHd_075 : 39.404 20.650 9.961 7.809
MuHd_095 : 40.366 22.856 10.162 8.716
MuHd_100 : 38.624 21.865 11.821 7.528
MuHd_105 : 43.831 24.409 13.019 8.449
Hmax_075 : 4.400 23.800 3.752 2.710
Hmax_095 : 9.450 9.188 -0.772 3.535
Hmax_100 : 2.000 6.500 -0.788 1.860
Hmax_105 : 4.667 3.400 -1.644 2.532
CLASS NOT-MITO MITO(/CHLORO)
DFM : 0.0084 0.9916
DFMC : 0.0022 0.9978
This protein is probably imported in mitochondria.
f(Ser) = 0.0962 f(Arg) = 0.0673 CMi = 0.58548
CMi is the Chloroplast/Mitochondria Index
It has been proposed by Von Heijne et al
(Eur J Biochem,1989, 180: 535-545)
-
Fasta :-
>PKNH_0827900
ATGCGGCTGCACGTGTCACTGCGGGTGGCATTGCCCCTGCACAGGAGCACCCTGAAGAAA
ACCAATCAACGGGCCCTACCCTACGTACAAACTAGGGCGCTCATGAACACCCCAAAAGTA
AGCACCCTCAAGAGCATGGACATGCCGAGTATCCGGAAAAGCGTACAGTTGAGCAAAAAA
AAAAATAATAAATCAAAAAATATATGGAGAGATTACCTGTACGTAGTTATTTTCAAACTT
TATAACAACCAAGTCGGGAATTATGTGGATCTTATGATAATTTTAAAATTGTTAAGCAAA
CAAATCTCTTTGGAAAAAAAAGTTCTGAATTTTATCTGTGAGAAACTGGAGAATCACATG
TACAGACTGACCATAAGGGAATTAAGTCTACTAATGTTGGTTCTCAGAAAACACAAGTTC
GATAATTTGTATTTCCTGAACTTGCTAGCCAAGTCTATACTAATTAAAATGAATAAAAAT
TTGTCTTACAAAGACTTAGCATTGATAACCTTCAGTCTATCGAGAAGCATTTCCTTGACA
GAGAAAGTATACACGAATGAAATTTTCCATCTAAGTGTTGTAAAAATACACAACGATTTG
AAGAACCTAAATTTCCACTCCCTCACTTTGTTCTTCTACTCCTACAGTCTTTATTTTCTT
CACCATTTTGAACACTTCAATTCCTTTTTTTATATGACCAAGAATTTTATCCAAATGATA
AAAAAAAATATGTATCTCTTCAACCCCACTGATTTAATGTTTACCTTTTTATCTGCCATC
CATATTTACGACTCTTTCAATAACGGTGTAAGGAATGTGCAATGCGACAGCGTGAGGATT
TCATCAGGTGCAGAAGTTTCACTTACATATGCAACCTTACCCAGGGGAGAACCTACGAAT
AATTATGCATTAACTAATTATACTGAGACTTATGATCAACCAACACTTGTACCCTCAAAG
ATGACACGCGCAGAAAACCCCGCCACGATGAGTGCTACCACAGAGGCACTTCATCAAGCA
AATTACCCCAACGAAAAAGTGGAGAAACAGGATGGAAAAGCCCGCGAAGAGACACTTTCT
AATTTCCTAACCCAAGTGAGGTACGCAATTGTAGAGAAGCTAAAGTGCTTCAAAGTGGAA
GAAGTGACAAACGTACTGTTTGCCTCGATAGATAGAGATTTCACCATTAACAAGAGGTAC
TTCCCCTTTTTGCAAAAGGATAAAATACACATTGAGGATTACCTGCATGTGTACGTGAAG
AAGGGTCAGATGTGGGAAGAGCAGCATGTGGAAAGGGGAGAACTCCCACATTGTCGTGCC
AATCCCCCCGCGGAAGAATCCTCCCATGTACCCCCCCAACCGACAAACTATATCATCAGA
ATTAGCCAAGGCAATCACCAGGAGGAATCATTCATTCATGCACTGTCGGAGGAAATAATT
TATCGTAACGAATTCCTGACAGCGCGACAAATACTTTTATTGCTCTACGCTTTGAGGGGA
AATAACACCCCATTTATACAATTGGAGAAAATTCTATTGAAGCGCTTGGTGTGTCTGGAA
AAGGAACTAACGGTCAGTCAAGTTATGTTCGTCTATCAGTACCTCTACTTCAGGACATGC
TTGTTTCACGAACTGAGGAGGTATGCTCCAAAAGTGTATAAACATGATAGGGGAAAAAAG
GAGGTGTTCCTCTTATGCGATGGTGACAGTGCAACCGGAGAAATTAGCCAACAGGAAGAC
CAATCTACCTTGCCAAGTGAACGAAAAAATGAGAAGGACAACAAGGTGTACTACCTCCAC
GATGATACATCCCACTTGAAGAAAAGGAGAAAAAGGGATGGGACAAAAATTTTCCTGTAT
GATAATTTTATTTTTAAGTACCCAGCACAGGTCGTCAGTACAGAAGGGGAAGAAGTAGCA
AATTTACAAGAAAAAAAAGAATTAAAAAATTTCATTAGTATGAAAAGAAGGAGAGATTCC
TTATCAGAGGTAAATCGTTCAACCGTAAGGAATGACGAGGATAAGATACTCACTGGCGAG
ACCGAATACCAAATCACTTGTTACAATTTCTACCTGGATGTATACAAAATTGTTTGTTTG
AAAATGTTAAACTTCCTAAAAAATGAAAACATAAACACCTGCACAATATTGCATGTTCTG
GATATATTCAAAAAGTTAAACATAAGAGACTATAGAATGATCAGACATCTGTATAAAATG
GAGAAAGATATTTTATTTTTGGATAATGCCAACCTGGAGAAGGTGTTAAAAATCCTCCTT
AATTTTAACCTCTATAATATAATGCATTATTTGGGGGTGCATAGAATTCTGCAATTTATT
AATTTCAATTCCGTGGATGAATGCGTGTCCATTTTGGGTATCCTTGGGATGATGTCCACG
CGGAAGAAGACGGGGCACCATGAGGGGAGGGAGAAGGCATTCCCTAATTTTCCTACCGCA
TGCACTGAAAATGCTATCAATTATATTTTAAAAAATTTAAAAGACTTGTCCAATATTAAT
GCGTTGGAGAAGATTCAGATAACGCCCGTGTACAATTCCTTACCATTGAAATATTTCAAG
CTTTTCAAAAATGTGCGCTCTGTGAGAGGTTTGGCTCAACTTTGGTTTCAACAAAATGGA
GGGAAAAAAAATCACCTTGCCTGGTCAGGTGAAAATGATCCAAATAGGATAAGAAACTTT
GTGTTGCATTTTGTGGATGACGTTAATTTGGGAAAAGTGCCATTTGGGTCAAATGGTTCT
TGTCGACTCGACTACGGGGGTGCCAATCAAATGAGGCAAACTCCAAATGCCACTGCCCAG
GTGAATTGCATCACCGAGTACAGAGATATCGAGGAGGAAATATTTCAGGATCTGCACACA
ATTTTTCCGCAGACAGTTGAAGTTTATAAAAATGTGAATGTGTCGAATTACACATTTCCG
ATGGCCATTAATTTGCTAACGCTGGGGGGTGCAATACCTAATAGATCTAGTCAGGCTAAC
GCGACTGACTTGGTAAAGAAACACTCCCTTGGAAATCAATGCGGAAGTGATCGACCCATG
CGGATACAAGAACTGGATAGGAAAAATACCCTCCTTGTGGACATTCTGTATGGCCAGGAT
TTTTACTACTGCCTGAGTGGAAACAAAGAGCAACAGTTGAAGAAGCACAACATTTACGTG
GCCTACTATGGCATGCATGCGAATAAGACAAATAAGAAAATGGTTAATTATAAAAAGTAC
AGCATATTGAAGTGCATTAGAAAAAGTGGGTACCATTTCATTTGCATTGATGCCAAGAGG
TATGTAAAAAACAAAAAAAAAAACCGAGACAATCACCTGAACAGAGATTACCTAACTAGT
TTAATACTTGATTTGATTAAGAGGAAGAAAAGAATTCCCCTGGGTATAAACAGAAGGGTT
AGTCGAAAAAATACCTCAGGTACATCATCGCGAAAACGGCGCAGAAGAAGGCCACTTTTG
AGAGAAAAAAGGCACACGTGCAGCGTTCAAGGGGTGATTTCCCATGTAGAAACAAACAAA
TACCTCCACAAGGTACACACTGGGGGGGAAAAGGTGAGCTTCCCCGGGACTAACCATCTG
GAGAAGTTGCGGTACATGTTCCAGCTTGGATAA
- Download Fasta
|
ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method |
|
PKNH_0827900 | 281 S | SVRISSGAE | 0.991 | unsp | PKNH_0827900 | 281 S | SVRISSGAE | 0.991 | unsp | PKNH_0827900 | 281 S | SVRISSGAE | 0.991 | unsp | PKNH_0827900 | 660 S | RRRDSLSEV | 0.997 | unsp | PKNH_0827900 | 1141 S | NRRVSRKNT | 0.998 | unsp | PKNH_0827900 | 1150 S | SGTSSRKRR | 0.996 | unsp | PKNH_0827900 | 124 T | MYRLTIREL | 0.992 | unsp | PKNH_0827900 | 162 S | NKNLSYKDL | 0.998 | unsp |


