

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PKNH_0835300 | SP | 0.257902 | 0.604029 | 0.138069 | CS pos: 20-21. THA-KR. Pr: 0.9742 |
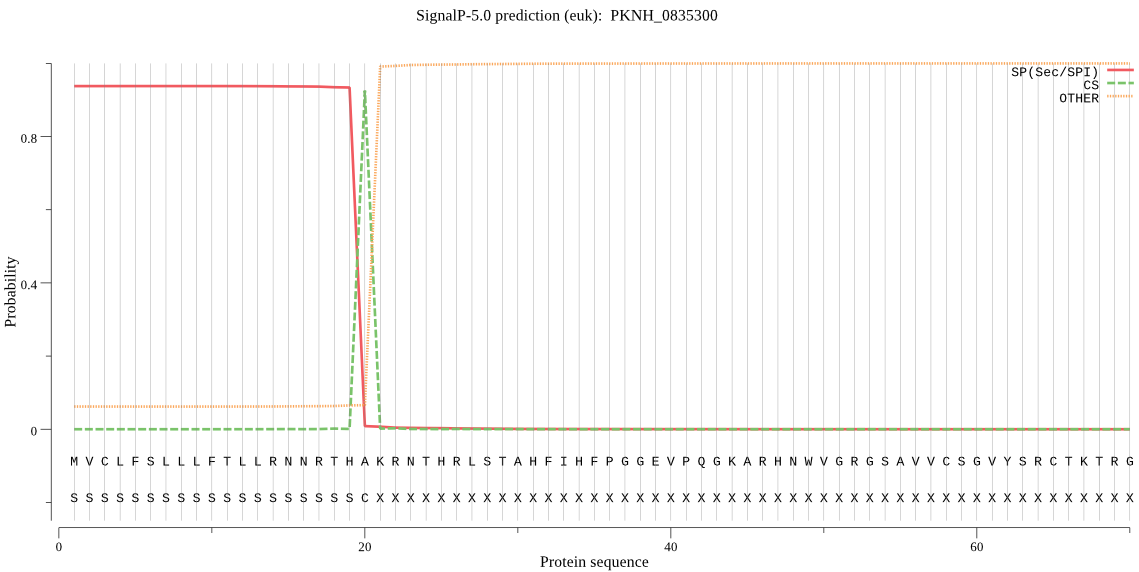
MVCLFSLLLFTLLRNNRTHAKRNTHRLSTAHFIHFPGGEVPQGKARHNWVGRGSAVVCSG VYSRCTKTRGLSSNSLLRSIEDEEENATLRDEGASDGASYITTDGVARSDPGTKEMKSVK NGNLCMDAAKGVGGFLGGEGRHLEEISPEGKPIHKGRSRMSRRYRRREGIIPRVEDIKDM KKDLKLFFFKKRIIYLTEEINKKTTDEMISQLLYLDNINHDDIKIYINSPGGSINEGLAI LDIFNYIKSDIQTISFGLVASMASVILASGKKGKRKSLPNCRIMIHQPLGNAFGQPQDIE IQTKEILYLKKLLYNYLSTFTNQTTETIEKDSDRDHYMNALEAKRYGIIDEVIETKLPHP YFDEAVRESSSG
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PKNH_0835300 | 161 S | RSRMSRRYR | 0.996 | unsp | PKNH_0835300 | 161 S | RSRMSRRYR | 0.996 | unsp | PKNH_0835300 | 161 S | RSRMSRRYR | 0.996 | unsp | PKNH_0835300 | 332 S | IEKDSDRDH | 0.998 | unsp | PKNH_0835300 | 79 S | SLLRSIEDE | 0.996 | unsp | PKNH_0835300 | 118 S | KEMKSVKNG | 0.996 | unsp |