

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PKNH_0913000 | OTHER | 0.999288 | 0.000671 | 0.000041 |
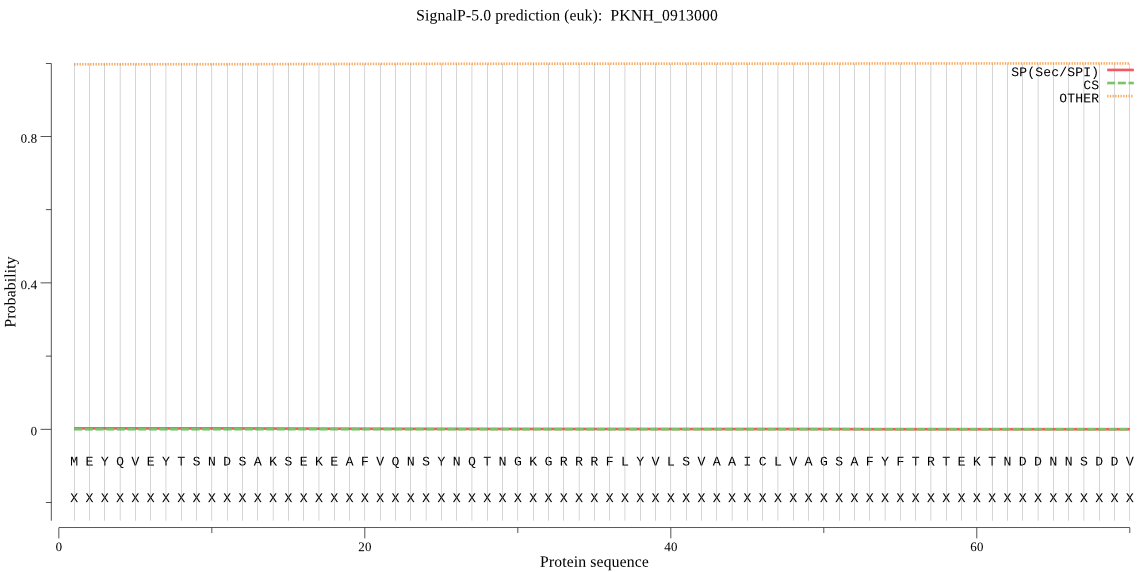
MEYQVEYTSNDSAKSEKEAFVQNSYNQTNGKGRRRFLYVLSVAAICLVAGSAFYFTRTEK TNDDNNSDDVIINTLLKSPGGKKFIISKLTELVASYDKEGNLQEQKPSNELISFVDRNEK SEENFRKRFGNLKNGKKIVDINFADSRFLMTNLENVNSFYLFIKEHGKKYQTPDEMQQRY LSFVENLAKINAHNNKENVSYKKGMNRFGDMSFEEFEKKYLTLKTFDFKSNGLKSTQLIS YDDVINRYKPKDDKFDHTKYDWRLHRGVTPVKDQGDCGSCWAFSTVGVVESQYLIRKNEL VSISEQQMVDCSLQNNGCDGGFIPRALEDIIEMKGLCSTEAYPYVGEVPEKCKYDMCDRK YKINSFFEIPEFKFKEAVRYLGPISVNIAVSDDFAFYQGGIFNGECGRTTNHAVILVGFG AEDVYDSDMNTTRKRYYYIIKNSWGVSWGERGFIRMETDINGYRKPCLLGLEAFGVLVE
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PKNH_0913000 | 212 S | FGDMSFEEF | 0.995 | unsp | PKNH_0913000 | 212 S | FGDMSFEEF | 0.995 | unsp | PKNH_0913000 | 212 S | FGDMSFEEF | 0.995 | unsp | PKNH_0913000 | 12 S | TSNDSAKSE | 0.993 | unsp | PKNH_0913000 | 15 S | DSAKSEKEA | 0.998 | unsp |