

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PKNH_0913700 | SP | 0.003463 | 0.996535 | 0.000002 | CS pos: 24-25. AES-FR. Pr: 0.8507 |
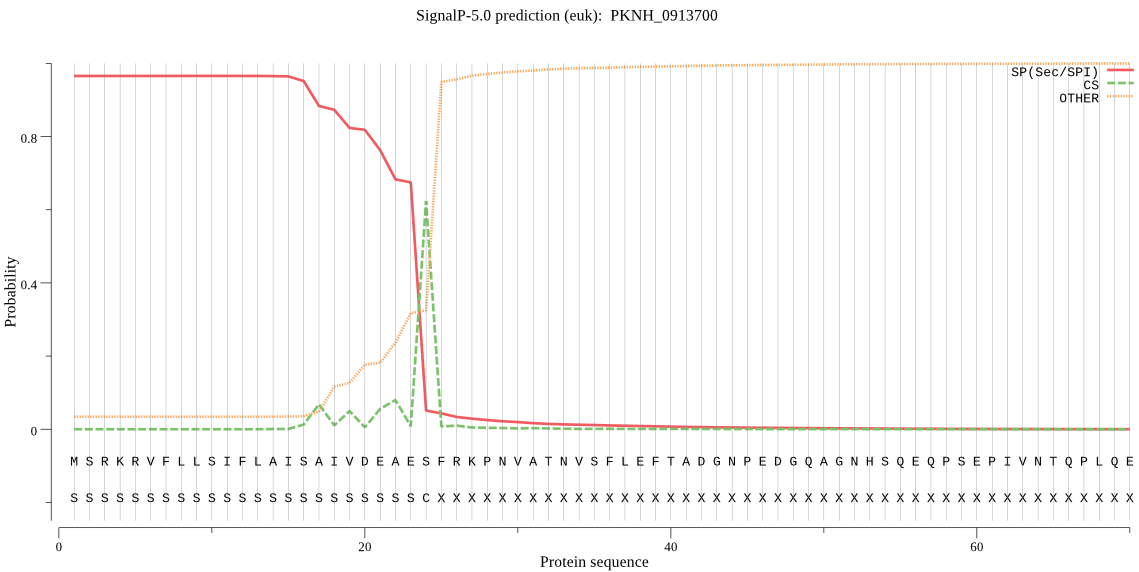
MSRKRVFLLSIFLAISAIVDEAESFRKPNVATNVSFLEFTADGNPEDGQAGNHSQEQPSE PIVNTQPLQENVSEEANHTEESTGGSAEKGNTGSQVEPTHGGGNEETPHQGEHDKGSNGN GTNVGASVDTSGKVDGTANGHAEEASNEHAVGNVGIEKNDTHNESTTTDSNSDNGQMDLH TSTPSDKEENSSPSMNNMHNTVDLKGENAFESTTPGEYKASQQNMPHEVVAKNDGEERDG GASNMGALPHLHDQHEQQGGDAKHTEHGEQVEHSDHAEKEEHAQHTEHAEHAEHSEHAEH AEHAEHADHAEKEEHAKHTEHGEHADHAEHREHADHAKHGEYADHADHSEHADHVDRSEH ADNVDHSEHADHTEEGEHQIYGTIHPEVETSVLSSLREKGIHVSSIERVILEIIESAKEG VKGLLKLRTSKDSGKLLQEALEKHSINQRDIQINNNIISLDMYDRILSEMFKILTDMSFY KDGHFYETLGLNKSILNQSLKEIKIKMLRTIGIPYAKLPPIVKNKEKGGTCAVNNLIISI TSKELAQRMAIMFSKWLAPEEYGSVVDLDKSIELNVLCSGAPILVQQWKYYQNMLGFEVG NEHAFLNLIDDLLVIDKRYSNNEEYSKVLKKIKKSKAFNYCTKVMRIAGNISSIPFNHEN NKTPSYSIIGSLGNLVKAHMGNYYTAIANRLNSYFLYAEKRNKKNSPLKVTSVCTLLHLT DMLFNCSDEHMKNILDLNTLKLNILNMQGKRVLQPLVKMNFLNETKNAVLKEICEPSNRL VDETLSKLLNMLSTGSHELLAAEVEKRGFDEDYIQEEIKNINESDKNIRDKGEDEVENFI FEDL
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PKNH_0913700 | 499 S | ILNQSLKEI | 0.991 | unsp | PKNH_0913700 | 499 S | ILNQSLKEI | 0.991 | unsp | PKNH_0913700 | 499 S | ILNQSLKEI | 0.991 | unsp | PKNH_0913700 | 564 S | EEYGSVVDL | 0.993 | unsp | PKNH_0913700 | 620 S | DKRYSNNEE | 0.995 | unsp | PKNH_0913700 | 824 S | NINESDKNI | 0.993 | unsp | PKNH_0913700 | 395 S | SVLSSLREK | 0.993 | unsp | PKNH_0913700 | 430 S | KLRTSKDSG | 0.992 | unsp |