

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PKNH_0914400 | SP | 0.002018 | 0.997941 | 0.000041 | CS pos: 25-26. TTA-DI. Pr: 0.9575 |
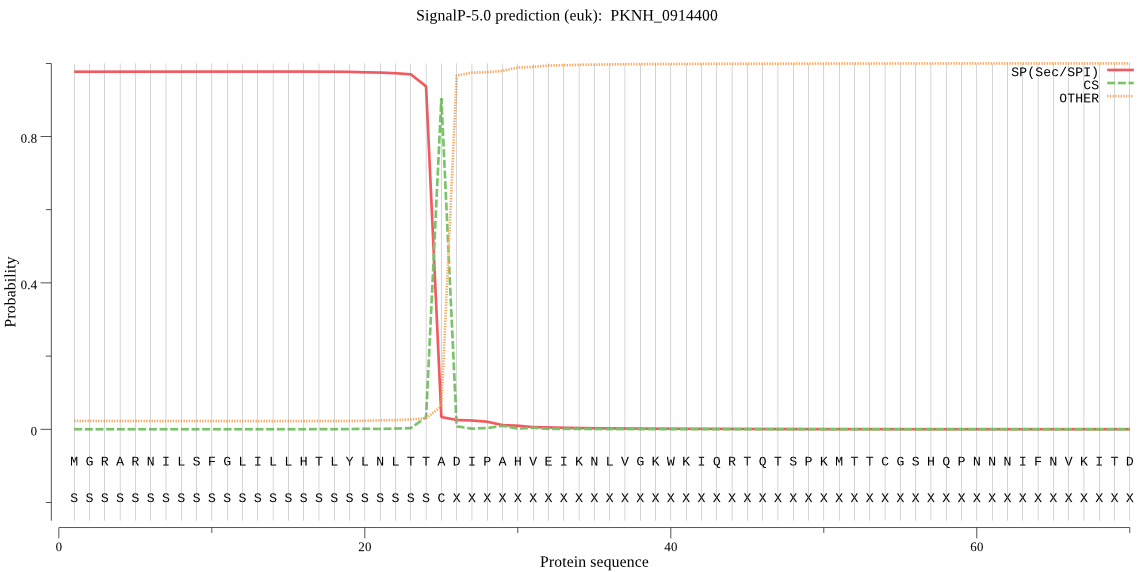
MGRARNILSFGLILLHTLYLNLTTADIPAHVEIKNLVGKWKIQRTQTSPKMTTCGSHQPN NNIFNVKITDYKKYLLDNHYKFTSDLYVILSDDFVPYGDMHDTTGNEHRKNWKVLVVYDE HKRKIGTWTTICDEGFEIRLGNETYTAFMHYEPTGKCGEAKEDDQTDSNGETDCYTTNYN KIRFGWVDINKSKDEKLYGCFYAERYHVSADPSDIASTQQNISNEASVDSFLNSHNFLYP ATSAAMSGSNDKPTFTKRKNMHIDQNSELYWHKMKHHGKKKPISREMMMNAKQRYACPCN KDEHVQDEVNNGDNPDQPVSPVSPVSLMQLGNRTGDDETNEMDLENYEDTEKSPHRELEI DELPKNFTWGDPFNKNTREYDVTNQLLCGSCYIASQMYVFKRRIEIGLTKNLDKKYLNNF DDLLSIQTVLSCSFYDQGCNGGYPYLVAKMAKLQGIPLDKVFPYTATEQTCPYKVDQFAM VTPAAGSNPSTATTKNLRQINAVMFSTGTRNDMHANFKNPISDDPEKWYAKDYNYIGGCY GCNQCNGEKIMMNEIYRNGPIVASFEATPDFYDYADGVYYVKNFPHARKCTVDATKKNFV YNVTGWEKVNHAIVLVGWGEEEIDGTMYKYWIGRNSWGKNWGKEGYFKIIRGLNFSGIES QTLFIEPDFTRGAGKILLEKLKNE
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PKNH_0914400 | 247 S | SAAMSGSND | 0.994 | unsp | PKNH_0914400 | 247 S | SAAMSGSND | 0.994 | unsp | PKNH_0914400 | 247 S | SAAMSGSND | 0.994 | unsp | PKNH_0914400 | 284 S | KKPISREMM | 0.992 | unsp | PKNH_0914400 | 48 S | RTQTSPKMT | 0.996 | unsp | PKNH_0914400 | 168 S | DQTDSNGET | 0.992 | unsp |