

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PKNH_0914500 | SP | 0.027777 | 0.970071 | 0.002152 | CS pos: 26-27. ASC-SA. Pr: 0.8101 |
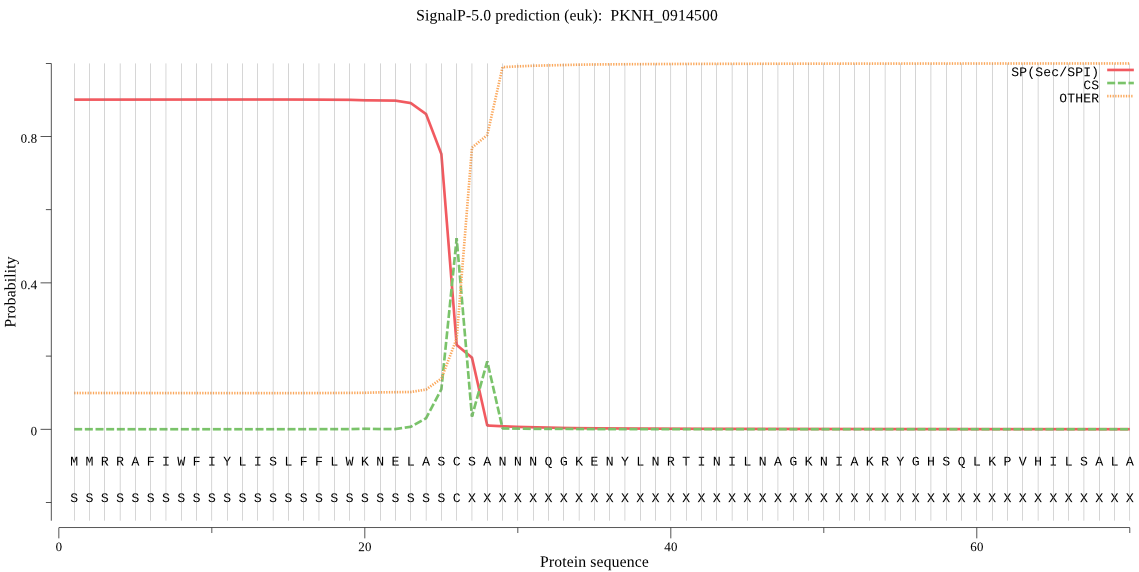
MMRRAFIWFIYLISLFFLWKNELASCSANNNQGKENYLNRTINILNAGKNIAKRYGHSQL KPVHILSALAKSDYGSNLLKENSVNASNLKQYLDTALEQTRAGPPLDNKSKIAYSDQVKE VLAEAEALANKYKSQKVDVEHLLSGLMNDELVNEILNEVYLTDEAVKGILKSKLEKTKKD KDGKSGGLYLEQFGSNLNEKVRNGKLQGIYGRDEEIRAVIESLLRYNKNSPVLVGQPGTG KTTIVEGLVYRIEKGDVPKELRGYTVISLNFRKFTSGTSYRGEFETRMKNIIKELKNKKN KIILFVDEIHLLLGAGKAEGGTDAANLLKPVLSKGEIKLIGATTIAEYRKFIESCSAFER RFEKILVEPPSVENTIKILRSLKSKYESFYGIHITDKALVAAAKVSDRFIKDRYLPDKAI DLLNKACSFLQVQLSGKPRIIDVTEREIERLAYEISTLEMDVDKVSKRKYNNLIKEFENK KDLLRTYYEEYVISGERLKRKKEAEKRLNELKELAQNYISANKEPPIELQNSLKDAQEKY MEVYKETLAYVEAKTHNAMNVDAVYQEHVSYIYLRDSGMPLGSLSFESSKGALKLYNSLS KSIIGNEDIIKSLSDAVVKAATGMKDPEKPIGTFLFLGPTGVGKTELAKTLAIELFSSKD NLIRVNMSEFTEAHSVSKITGSPPGYVGFSDSGQLTEAVRERPHSVVLFDELEKAHPDVF KVLLQILGDGYINDNHRRNIDFSNTIIIMTSNLGAELFKKKLFFDANNSDTPEYKRVFDD LRIQLIKKCKKVFKPEFVNRIDKIGIFEPLSKKNLREIVKLRFKKLEKRLEEKNIHVSVS EKAVDYIIDQSYDPELGARPTLIFIESVIMTKFAIMYLKKELVDDMDVHVDFNKAANNLV INLTAV
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PKNH_0914500 | 371 S | VEPPSVENT | 0.991 | unsp | PKNH_0914500 | 371 S | VEPPSVENT | 0.991 | unsp | PKNH_0914500 | 371 S | VEPPSVENT | 0.991 | unsp | PKNH_0914500 | 384 S | RSLKSKYES | 0.992 | unsp | PKNH_0914500 | 532 S | ELQNSLKDA | 0.997 | unsp | PKNH_0914500 | 657 S | IELFSSKDN | 0.996 | unsp | PKNH_0914500 | 705 S | ERPHSVVLF | 0.994 | unsp | PKNH_0914500 | 811 S | FEPLSKKNL | 0.994 | unsp | PKNH_0914500 | 840 S | HVSVSEKAV | 0.991 | unsp | PKNH_0914500 | 134 S | NKYKSQKVD | 0.995 | unsp | PKNH_0914500 | 279 S | TSGTSYRGE | 0.992 | unsp |