

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PKNH_0924500 | OTHER | 0.870529 | 0.117783 | 0.011688 |
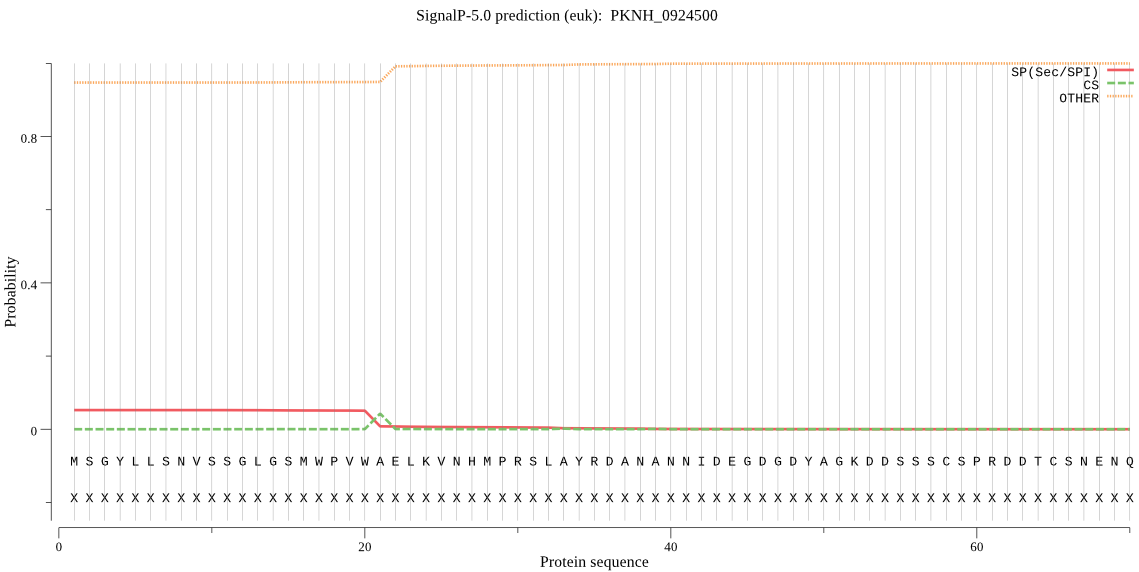
MSGYLLSNVSSGLGSMWPVWAELKVNHMPRSLAYRDANANNIDEGDGDYAGKDDSSSCSP RDDTCSNENQSNNQNDAKEEKPTLECSPCKFWKGRKKNNNLDSMEELLFKLTNGKFKAEK HHVYTADGYRLNLYRIVNTNEKENLSKKKEVFCLNHGLFESSISYTCKGYESLAFQIFAN DYDVWISNNRGNAFTKFVGKNYALKKLRERYSLQDLKDMGVDVSEEMDSNLSTDEGTEGE QQQQQHQNNDKDGDLVYGTEEGANPQGPFGESSDCDSKMVHSFPYVPRELENGDATEVAS EMTTEKTAEKIAEMTTEVETENAAKGEDEEEVEELVNLNKKKEGLHDMDDDDVNKQNEPE INNWTFEDMGTKDLPPIIKYIREKTQREKIVYVGFSQGSIQLLVSCCLNDYVNRSIKRTY LLSLPIILKTKGEMLMSVRMLLAISRWNKAVVGSKEFLQKVLPEKICNSLIINSADLLTR NVFKFFTDNVDDSYKRIYYRHTPSGTTSRENLRKWLTCPNHEPVSEAIDKYAYKCSFPIT LVYGINDCLVDAERSIEYMKKKFTKNDLKIITKPEWSHIDPILTDNENIVLSCILEDMKR EEEEKKQEGM
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PKNH_0924500 | 212 S | RERYSLQDL | 0.992 | unsp | PKNH_0924500 | 212 S | RERYSLQDL | 0.992 | unsp | PKNH_0924500 | 212 S | RERYSLQDL | 0.992 | unsp | PKNH_0924500 | 232 S | DSNLSTDEG | 0.993 | unsp | PKNH_0924500 | 385 T | IREKTQREK | 0.996 | unsp | PKNH_0924500 | 415 S | YVNRSIKRT | 0.995 | unsp | PKNH_0924500 | 493 S | NVDDSYKRI | 0.996 | unsp | PKNH_0924500 | 508 S | SGTTSRENL | 0.991 | unsp | PKNH_0924500 | 577 S | KPEWSHIDP | 0.994 | unsp | PKNH_0924500 | 59 S | SSSCSPRDD | 0.998 | unsp | PKNH_0924500 | 146 S | KENLSKKKE | 0.992 | unsp |