

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
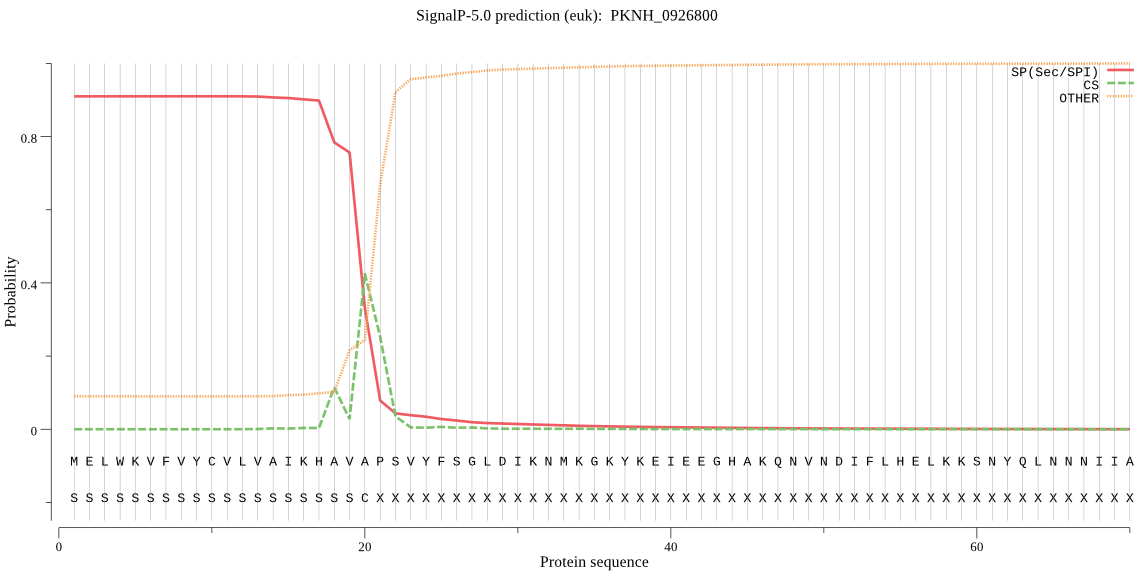
| PKNH_0926800 | SP | 0.028431 | 0.970193 | 0.001375 | CS pos: 18-19. KHA-VA. Pr: 0.3200 |
MELWKVFVYCVLVAIKHAVAPSVYFSGLDIKNMKGKYKEIEEGHAKQNVNDIFLHELKKS NYQLNNNIIALSTSRHYFNYRHTSNLLTAYKYLKNAGDNMDRNILLMVPFDQACNCRNMV GGTIFNEYEKPSSEDLKEKKMKENLYSHLNIDYKNDNIRDEQIRRVIRHRYDALTPVKYR LYTNGNREKNLFIYMTGHGGVSFFKIQDFNIVSSAEFSLYIQELLIKNIYKYIFVIIDTC QGYSFYDQILNFLEKNKINNVFLMSSSDKNENSYSLHSSRYLSVSTVDRFTFYFFSYLEN INRIYADEPYKNTKAFSLYNILNYLKTQHLISTPTINNFKFSVSMFMHSKNILFYDSSGF YLSKDTQEGALDGLGKSSYNAKHKTDKTATRNTCLGDLSACGHMKNEIHTHMGNLYSRSS FYNNVEVYFKEESHFTDYYFTAECFSGGYLLPSFLFLVIIALCLLFFLFT
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PKNH_0926800 | 420 S | YSRSSFYNN | 0.992 | unsp | PKNH_0926800 | 420 S | YSRSSFYNN | 0.992 | unsp | PKNH_0926800 | 420 S | YSRSSFYNN | 0.992 | unsp | PKNH_0926800 | 132 S | YEKPSSEDL | 0.997 | unsp | PKNH_0926800 | 285 S | YLSVSTVDR | 0.992 | unsp |