

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PKNH_0940900 | SP | 0.225326 | 0.672180 | 0.102494 | CS pos: 21-22. CRC-FF. Pr: 0.7828 |
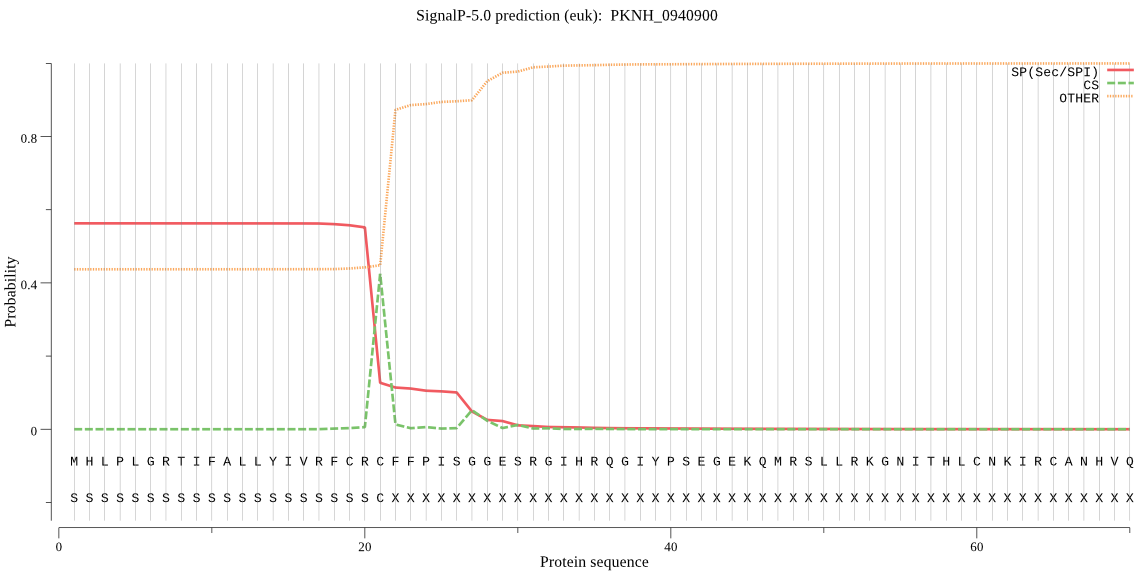
MHLPLGRTIFALLYIVRFCRCFFPISGGESRGIHRQGIYPSEGEKQMRSLLRKGNITHLC NKIRCANHVQRYEGRRGIFSSTGSSGKRNPLSSRLFSSFETKTPGSQVEQVVDGIAVTIY DNTNIFKEEISNTPIVLIHGCYGSKNNFRVFSKSLKSNKIVTLDLRNHGNSKHTDSMKYE EMESDIKKVLDELHIRKCCLVGFSLGGKVSMYCALKNQSLFSHLVVMDILPFDYNEKKYH VKLPYNISHMTKILFNIKTKLRPRNKAQFLAHLRAQVPDISSSFEQFICTSLKEERAKVG KATGEVDMPIGGTDLIRDSSMGTEGAIRYSENPATEQKNLVWKINVDTIFRELPHILSFP LNHQEHKYHNPCSFIIGTKSDLVYTMPQYESIIENYFPSSQKFILPDATHTVYIDNAKEC ADIVNRTLRL
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PKNH_0940900 | 106 S | KTPGSQVEQ | 0.996 | unsp | PKNH_0940900 | 106 S | KTPGSQVEQ | 0.996 | unsp | PKNH_0940900 | 106 S | KTPGSQVEQ | 0.996 | unsp | PKNH_0940900 | 154 S | VFSKSLKSN | 0.995 | unsp | PKNH_0940900 | 176 S | KHTDSMKYE | 0.993 | unsp | PKNH_0940900 | 319 S | LIRDSSMGT | 0.992 | unsp | PKNH_0940900 | 85 S | STGSSGKRN | 0.996 | unsp | PKNH_0940900 | 97 S | SRLFSSFET | 0.996 | unsp |