

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
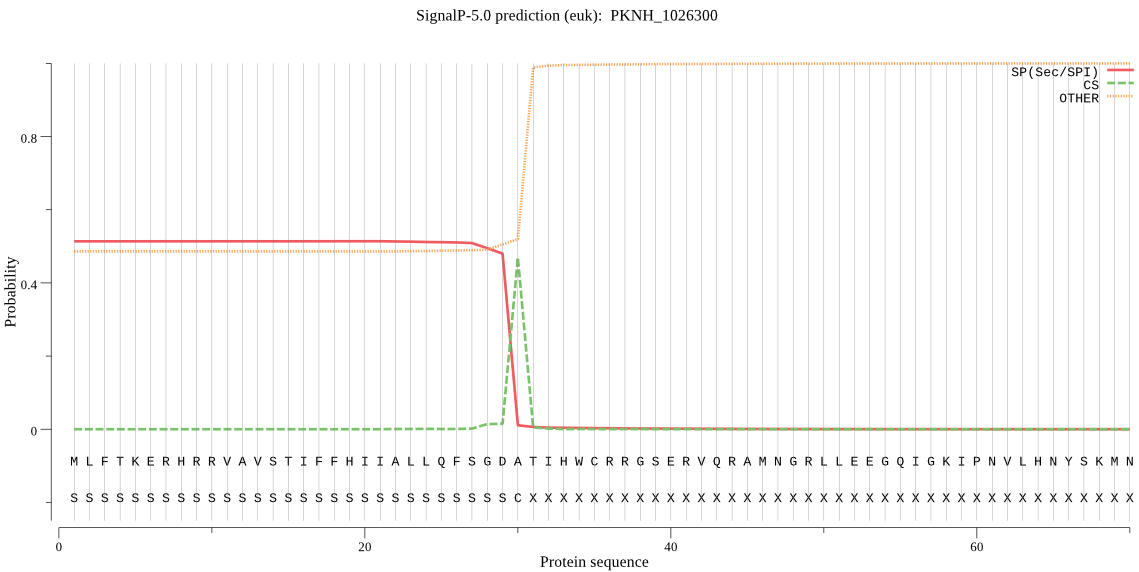
| PKNH_1026300 | SP | 0.437167 | 0.546011 | 0.016822 | CS pos: 30-31. GDA-TI. Pr: 0.7047 |
MLFTKERHRRVAVSTIFFHIIALLQFSGDATIHWCRRGSERVQRAMNGRLLEEGQIGKIP NVLHNYSKMNLSTKSFVQVKVKDSHDRNIRRGGVTTSEGNYHIPNCDDTDESSNFLKKFC RSMKSFVGITPHDKDWCLKYEKFRRFKQHFVYMVQNNLSLGKTRVCLIDTGLDLQDKVVR QFVKIYRWEHSKEGDNPSERSGGGEYDTHLQLEERDDEGASPSWAYPKGGDAKYDGINTE RCNEENYARCQSSDIDDVDMHGTFIANTVIRRDLLMKREMYKKNVELIVCKAFGDREETN SHLMPLIKCLEHCKGSGAKVIHVGYNVEGESEKLVEVMQELERAQIVVVSPSLRVYTGQS DESQQPRKEHGEEHSTEKLYPSSFADTFENVFSVGALRNSTQGGFVPISGNGNPKGEKQK WKVLHKRENTTLFSFSYGKTFPFGRSPSSMVEDGEGYASADFVNILVMILNVIPKLSIRR MRHILKRSIVKRSEMKGLSKWGGYIDPLKVIDATLKERNELCKTFFGELDLDLEAEGGSS SFRGDLSKGGFSGGSMTEDLGDWVEKPTTGTEEQIFDKTITGDVTPRLNDEQEESEKATG HLEDQVIFPQQRGEMEEMERHLVGETLNGLDPEERNDLGRYDMAHYDMDVISKWEEDFSD GVAALREDPASDTTFTSMYNYEENVYVPDEAKESFYEDAGRVTVTGEMSSLPLGFSFLEN HTNDRGSDLPLYRTNERGQVYASGDGTPGLPLNRRSQDENGFPERWGQQGRQAMIEEDSG EGYHHGQDDWVTQMKGTKSVDDPEYDVGRMYRDSPEGIPAEELLRSGWRGTTPPLSRWNR NGETSLWGDENEGYVFSPNVDNNWGEKDGRNRRNPQRRRSRDDLERRRRRFPRAMPRQRR RIPRRGRSKRRLKQREAGVVRKNGRNGKRRNENGMRSRREGEYSNMPMRRRPVRGKPVPG FAPKMPRVVMGRR
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PKNH_1026300 | 449 S | RSPSSMVED | 0.994 | unsp | PKNH_1026300 | 449 S | RSPSSMVED | 0.994 | unsp | PKNH_1026300 | 449 S | RSPSSMVED | 0.994 | unsp | PKNH_1026300 | 595 S | EQEESEKAT | 0.991 | unsp | PKNH_1026300 | 756 S | LNRRSQDEN | 0.993 | unsp | PKNH_1026300 | 799 S | KGTKSVDDP | 0.997 | unsp | PKNH_1026300 | 814 S | MYRDSPEGI | 0.997 | unsp | PKNH_1026300 | 854 Y | ENEGYVFSP | 0.99 | unsp | PKNH_1026300 | 880 S | QRRRSRDDL | 0.998 | unsp | PKNH_1026300 | 908 S | RRGRSKRRL | 0.997 | unsp | PKNH_1026300 | 937 S | NGMRSRREG | 0.997 | unsp | PKNH_1026300 | 943 Y | REGEYSNMP | 0.991 | unsp | PKNH_1026300 | 39 S | CRRGSERVQ | 0.994 | unsp | PKNH_1026300 | 253 S | RCQSSDIDD | 0.991 | unsp |