

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
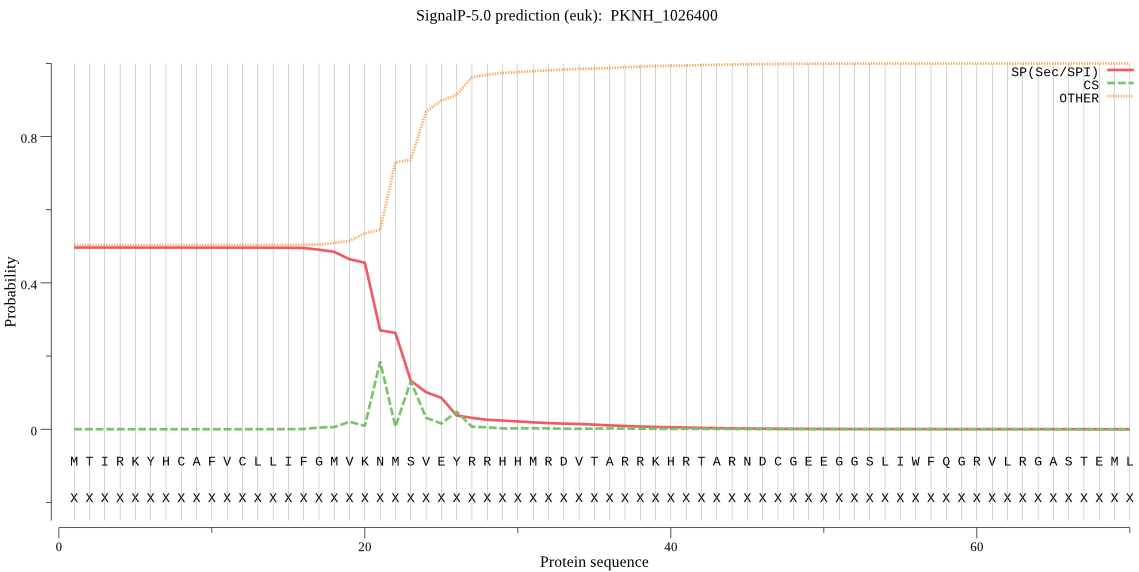
| PKNH_1026400 | SP | 0.475635 | 0.521307 | 0.003058 | CS pos: 21-22. VKN-MS. Pr: 0.4172 |
MTIRKYHCAFVCLLIFGMVKNMSVEYRRHHMRDVTARRKHRTARNDCGEEGGSLIWFQGR VLRGASTEMLRGEGLETGVHSSSMSSASPSSPPQVDHLLQDDSHNMDEMITKLREKKMII KFKQEENALQSALVEENTVNLLGSCGRVKKLNHIHLYLYEVFSNINERALKNCLQLLSSE HMLVEEDFQIHPVEGGGCGSGMDDLIPIKEMGGIRGEAPVRNGNSLRRHPGSDGSFQLSN QMAFNNFLKRLQSNRKGTNIIKGYDQTRMKEGTELSEPHEQNDVNVCIVDTGVDYNHRDL HDNVVHVLHGRDVILDQKENDENRSAEKECDRVDGDDGPDGMDNHGHGTFIAGIIAGNSK RENHGIKGICKRAKLTICKALNSKNAGFVSDILKCFNFCASKEAKIINASFASTKNYLSL FEALKTLEEKNILVVSSSGNCCPTSESKNAFPECNLDVKKVYPTAYSKKLRNLITVSNMI QKKNAQVSLSPDSCYSAKYVHLAAPGDNIISTFPKNRYAISSGSSFSAAVVTGLASLVLS INGSLRIEEVIRLLRESIVQTESLRNKVKWGGFLDVHHLVSSTIALSHGRTEIAKE
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PKNH_1026400 | 359 S | IAGNSKREN | 0.992 | unsp | PKNH_1026400 | 359 S | IAGNSKREN | 0.992 | unsp | PKNH_1026400 | 359 S | IAGNSKREN | 0.992 | unsp | PKNH_1026400 | 557 S | LLRESIVQT | 0.993 | unsp | PKNH_1026400 | 587 S | TIALSHGRT | 0.993 | unsp | PKNH_1026400 | 66 S | LRGASTEML | 0.991 | unsp | PKNH_1026400 | 88 S | MSSASPSSP | 0.994 | unsp |