

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PKNH_1113200 | SP | 0.002129 | 0.990507 | 0.007363 | CS pos: 18-19. GNA-FL. Pr: 0.9375 |
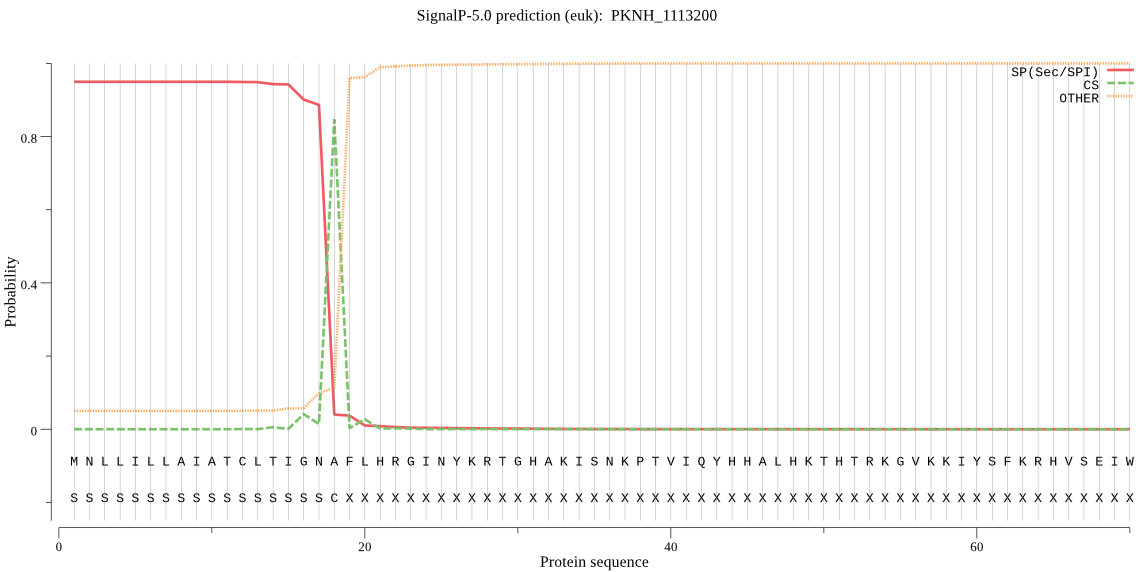
MNLLILLAIATCLTIGNAFLHRGINYKRTGHAKISNKPTVIQYHHALHKTHTRKGVKKIY SFKRHVSEIWNNIKTRRIENYMEDFFQTQQVKNTLQNNFLCFKHLFNKCKLDRVIIAVNV ALYFYLNRIDKDDEKKIFFHKGNLLEVKEQKAEKYKCNYHDVYKTKNFKTFFTSLFIHKN ILHLYFNMSSLISIYKIISQIYTNGQIFVTFILSGVLSNIISYICYVREKKENVLLKNLI LTNTSNENNFLVNKNNKIICGSSSCIYSLYGMHITYIIFFYFKHNYIVNSNFLYNFFYSF VSSLLLENVSHLNHVLGFFCGFFFSSLIVLFDRNA