

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
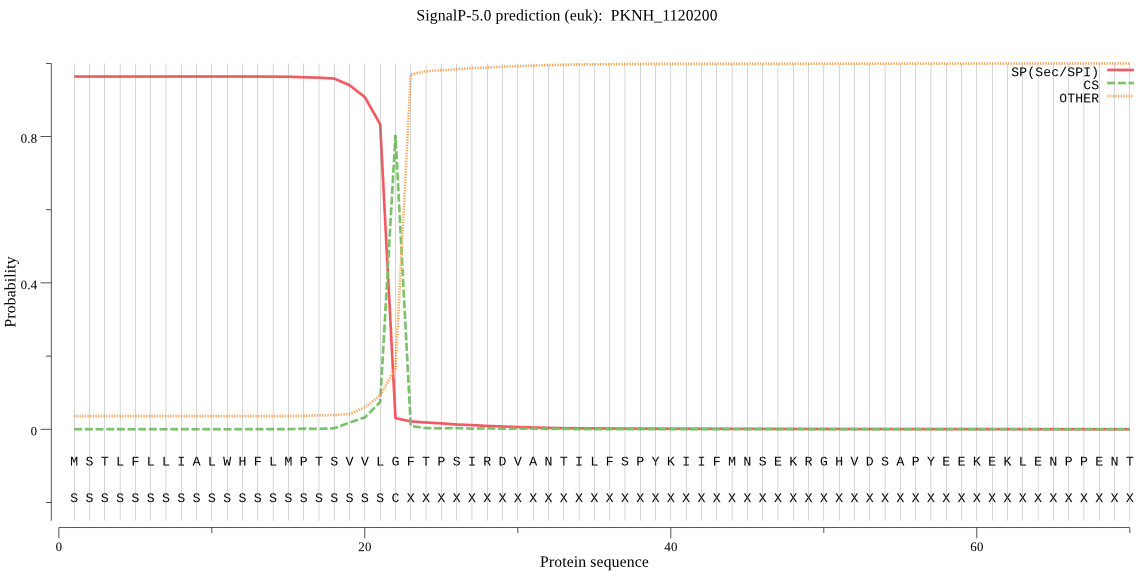
| PKNH_1120200 | SP | 0.026013 | 0.972815 | 0.001172 | CS pos: 22-23. VLG-FT. Pr: 0.8747 |
MSTLFLLIALWHFLMPTSVVLGFTPSIRDVANTILFSPYKIIFMNSEKRGHVDSAPYEEK EKLENPPENTEREEATVSVVEAEKEGKKTQGGLEEKKDEAPKAGVAKDNNVEEVAVTTED ASNGGKATKEEGVAKVEDVAVVEEVAKVEDVAAVEEVDKVEEAAEPAEEVIIVEEALKAT ANKVEPAEKEVTVEKEATVEKEATVEKEATVEKEATVEKEVTVEKEVTVEKAEGKSNEEA ARVDDTLKEEKYTFFWRRKPNKNKKKETKKKGSNDVSQEKDRKKLPTTFLLPGVGGSTLI AEYKDAMIHSCSSNLLNSKPFRIWISLTRLFSITSNVYCTFDTLRLVYDSEKKMYSNQPG VNITVEDYGHLKGIDYLDYINNTGIGVTKYYNTIASHFLSKGYVDGESIIGAPYDWRYPL YQQDYNLFKDTIEATYERRNGMKVNLVGHSLGGLFINYFLVHIVDKDWKQKYLNSVLYMS SPFKGTVKTIRALLHGNRDFVSFKIKKLIKLSISDSMMKAIGNSVGSLYDLIPYKEYYDH DQVVIIINTDSAPIDDNYMQSIVSECGIYNKDCYLNRTDVKLKVHTLSDWHVLLKDDLME KYNNHKQYRDRHFSMDHGIPIYCVYSTLKNKSTEYLLFSQKDNLNEELIVYYGIGDGTVP LESLEACNSFYNAKQKKHFENYSHIGILHSAEAVNYVYDSMQTTGGELDDAFDHVTAELT VEGSTEGGNVGGEELNKVPQQGKGNEEMANVPEIRVK
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PKNH_1120200 | 277 S | SNDVSQEKD | 0.99 | unsp | PKNH_1120200 | 277 S | SNDVSQEKD | 0.99 | unsp | PKNH_1120200 | 277 S | SNDVSQEKD | 0.99 | unsp | PKNH_1120200 | 527 S | NSVGSLYDL | 0.99 | unsp | PKNH_1120200 | 639 S | YLLFSQKDN | 0.996 | unsp | PKNH_1120200 | 70 T | PPENTEREE | 0.996 | unsp | PKNH_1120200 | 236 S | AEGKSNEEA | 0.995 | unsp |