

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PKNH_1215800 | SP | 0.019678 | 0.978907 | 0.001416 | CS pos: 21-22. VKS-AK. Pr: 0.9253 |
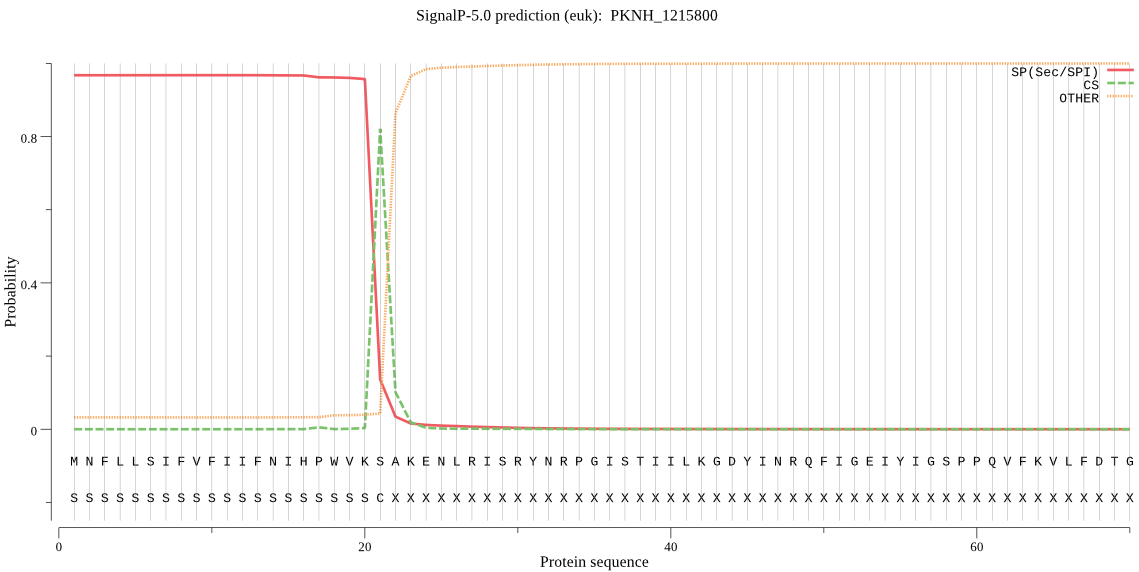
MNFLLSIFVFIIFNIHPWVKSAKENLRISRYNRPGISTIILKGDYINRQFIGEIYIGSPP QVFKVLFDTGSTNLWIPSKNCQTKGCRSKRKYDHRVSKNYKSVEKKNPVEVFFGTGKIQI AYVSDDVHLGDITVKNQEFGVASYISDDPFSDMKFDGLFGLGISDDKKRKTLIYENIPKG RSKKNVFSIYYPKNVDDDGAITFGGYDQKFIQPNENINWFDVSSRKYWTVKMIGLKINGV FLDVCSKNIGGYCEAVIDTGTSSIAGPKEDLILLTRLLNPRRTCHNRALLKRFSFLLSDE NGEQKEYELTPEDYIVNSFRVDPALKTPCNFAFMPINISSANRYLYILGQIFLQKYYAIF EKDKMKIGLAKSV