

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PKNH_1227700 | SP | 0.448149 | 0.548092 | 0.003760 | CS pos: 26-27. GTR-VN. Pr: 0.2131 |
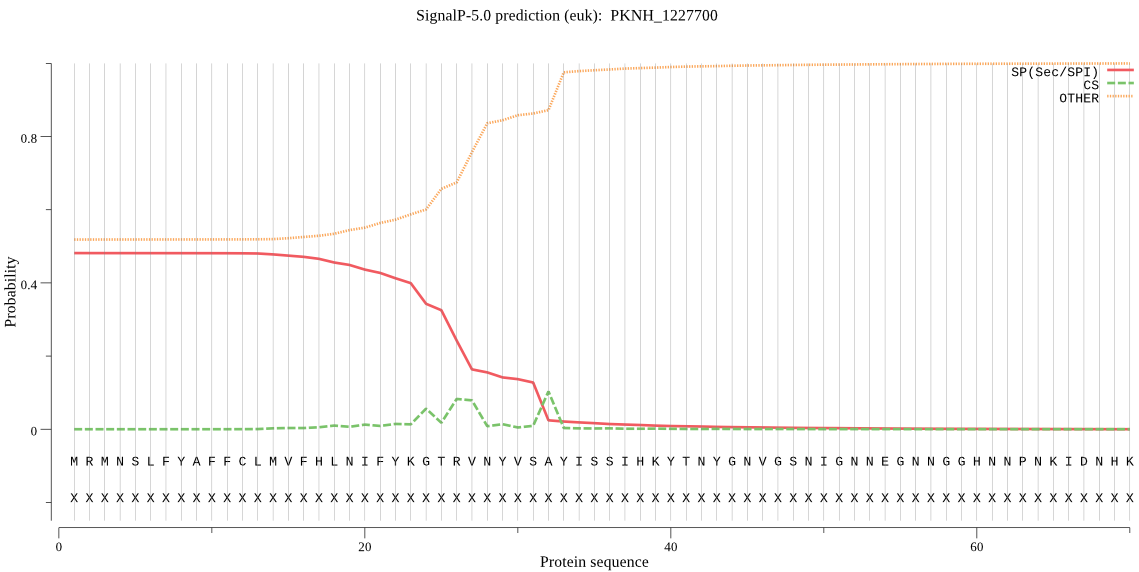
MRMNSLFYAFFCLMVFHLNIFYKGTRVNYVSAYISSIHKYTNYGNVGSNIGNNEGNNGGH NNPNKIDNHKISANETSNRSAASYFKKKFPQIYQQNGASVPSAVGDSTQTSEKKEFERTS QLQQEAEEPRQQLQESDNLEEDKDNMAAPIEERLANLKKVMQENNIDVYILINSDEHNSE IINDKDKKIYFLSNYSGADGILILTKDKQILYVNALYELQANKELNHDIFTLRVSRITNR DEIYETIASLEFNNIAVDGKNTSVNFYEKLKGKIESTYPEKKVEEKVIYGKNMNQIEKNE NINFLILEKSLVELKQNEVNNKQVFIHDRLYNGACAGQKLEKFRQAFSFDKKNVDKILLS ELDEIAYILNLRGFDYTFSPLFYGYLYFEFNREKDEFGKMILFTVSKNLSPSSIRHLNTI KVDVKEYETVVEYLRDNVSSKTMALTKAGNETSAVHTNLSEEFDSQKKYEISLSPYINLM IYMLFNKEKILLEKSPVVDLKAVKNDVEIDNMKEAHVLDALALLQFFHWCDEKRKTKELF NETEMSLKNKVDYFRSTKSNFISPSFATISASGPNAAVIHYEVTESTNSKITPSIYLLDS GGQYLHGTTDVTRTTHFGEPTAEEKKIYTLVLKGHLHLRKVIFASYTNSMALDFIARENL FKHFLDYNHGTGHGVGLFLNVHEGGCSIGPTAGTPLKPYMVLSNEPGFYLENKFGVRIEN MQFVISKKKTDNTEFYSFEDLTLYPYEKKLLDYSILTTEEIKDINEYHDNIRKTLLPRLK KNPSEYGEGVVKYLMDITQPISIN
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| PKNH_1227700 | 111 S | STQTSEKKE | 0.995 | unsp | PKNH_1227700 | 726 S | QFVISKKKT | 0.994 | unsp |