

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PKNH_1229800 | SP | 0.041176 | 0.958689 | 0.000135 | CS pos: 21-22. SKG-NV. Pr: 0.6646 |
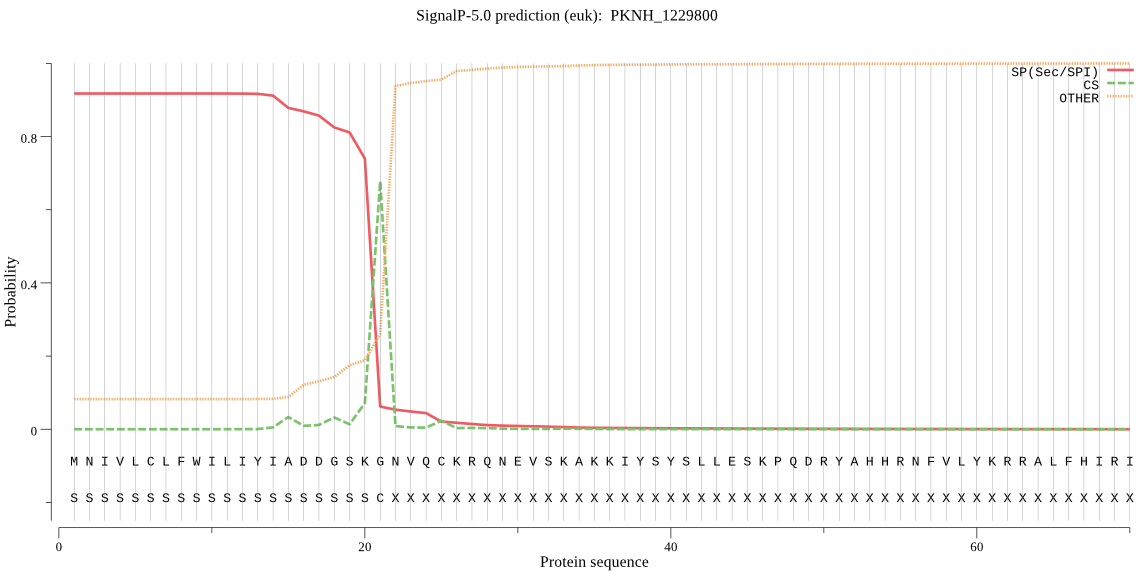
MNIVLCLFWILIYIADDGSKGNVQCKRQNEVSKAKKIYSYSLLESKPQDRYAHHRNFVLY KRRALFHIRIPRKNLERQNLFSNDLRYKKINKFLENKKANKLCYLHINNNNHGIKKKRKK NKTLQLFFEKKKLLQEYFLNLKSSFMDRYRNTKIITKLFLSSSLLMLTLNLIGLKPEDIA LHDKRIIRAFEFYRIVTSALFYGDISLYVLTNVYMLYVQSQELERSVGSSETLAFYLSQI SILSVICSYLKKPFYSTALLKSLLFVNCMLNPYQKSNLIFGINIYNIYLPYFSILIDILH AQNLKASLSGILGVTSGSIYYLLNIYAYQKFNKKFFLIPRFLRNYLDSLNIDDVL