

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PKNH_1269200 | OTHER | 0.994006 | 0.005954 | 0.000040 |
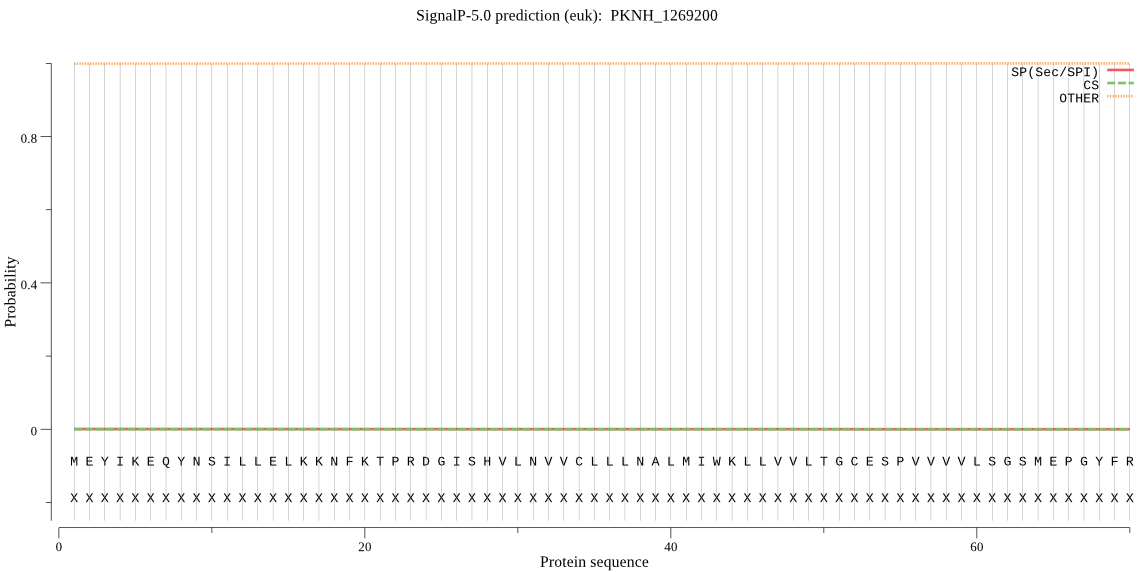
MEYIKEQYNSILLELKKNFKTPRDGISHVLNVVCLLLNALMIWKLLVVLTGCESPVVVVL SGSMEPGYFRGDTLALYHPPNIHAGDVVVYQINGRDIPIVHRILNIHISKDNKFHLLSKG DNNNIDDRGLYEFDQYWLENEHVLGLSVGYAPYIGMLTIWVNEYPAMKWGIVSLMLFMIL LGYE