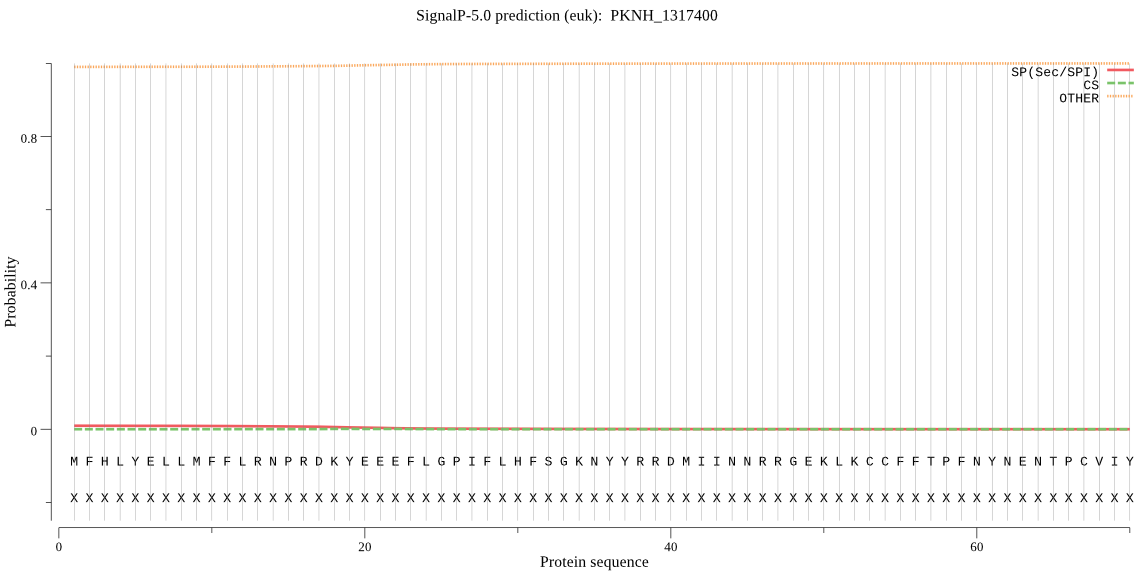
Fasta :-
MFHLYELLMFFLRNPRDKYEEEFLGPIFLHFSGKNYYRRDMIINNRRGEKLKCCFFTPFN
YNENTPCVIYTHSSCSCQLEALDILHILLICECSIFSYDCSGCGLSDGYFSTSGWNESQD
LFLILHHLRNVEHVKNFALWGKYSGAVSSIIAASLDANIKLLIVDSPYVSLTELYKTTFH
LSAKGKGEIIFKNICLYFAKRRIKKKFHYDIENICPIFFIEDVTIPTIYIISRNDKVVHP
AHTLYLAYKQKSEQKIIYTCEKASHAYESFSYDNKLTAAIRTVLFGAKKADVQNVFSVYT
YMRVFNGLMEKYSHEFCFIDSLIQKKLNRKDKIIDKVKQFICFKYQTKASLSSSTCFNQS
TIDSIRHTSTTSENDYMQYESSQNSLCWKCKSVPGEEHERDNHGLSEEQNQREDHGLVDE
YEYPFHDAHDFVNGQDATENPVNASALKGYDANKVPAFSPTFQHQGEQSGQRRFKVMHMQ
SARSSLKSKKNTFKKSLTWDTKLQSAVTYSKEDCPLELLKN