

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
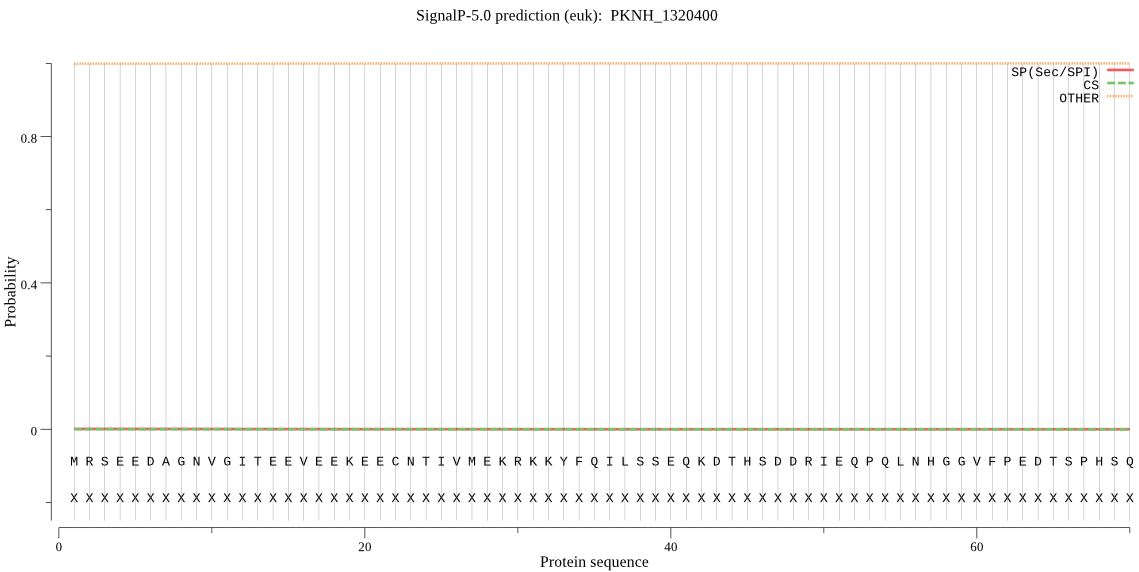
| PKNH_1320400 | OTHER | 0.999992 | 0.000002 | 0.000007 |
MRSEEDAGNVGITEEVEEKEECNTIVMEKRKKYFQILSSEQKDTHSDDRIEQPQLNHGGV FPEDTSPHSQEEEEERKKQKRDKKKDDSYPLLVVNKKGTIPKDDDRAKNQMLTTEEVINS PEETSRKGCLLKGERKYLLALKSIIRTRDGRHANGANGESGNHSGGCFDRGSDNHRRYSY FAKNTPRDETVNAEMHSHVLMDKSNGNRGSAWTPNVEKDPCVEASQGEKSEESARRKKWN FRRRIIGRIGHRAKGEDVCEEANKAAQPTIQSSTRSSTHHPANPSTEENNFVVYKVPRYL KKKNIKCPFVYRKVFFGKYGIINYDLKGNKKGTLVITFHGLNGTNLTFLDIQKTLVKYKY QVLNFDLYGHGLSACPKYNHRKKTYGINFFLTQTEELLTHLKLLHKDFYLIGFSMGCVIA ISFAKKYIKQVKKIILISPVGILEKKPFALKVLKLFPFLINISSFFMLPCFISKKNFKKA PKGASKKRQKSGNKEDPQCDTNDAYKDATDKSLEQDTQDDSDNDTSDYLYNRIMWQAFVK KNITHSILGCINNLKMWSAHDIFKEVGLHHIPVLILCGEKDNICSVHVFKNTSKFFINCH MIIFRNASHLVLVEKSKEINSCVLTFFHFPNNADLKTVHHMFPVDTLGNSVLCQRGDLL
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PKNH_1320400 | 120 S | EVINSPEET | 0.997 | unsp | PKNH_1320400 | 120 S | EVINSPEET | 0.997 | unsp | PKNH_1320400 | 120 S | EVINSPEET | 0.997 | unsp | PKNH_1320400 | 185 T | FAKNTPRDE | 0.992 | unsp | PKNH_1320400 | 230 S | QGEKSEESA | 0.996 | unsp | PKNH_1320400 | 233 S | KSEESARRK | 0.991 | unsp | PKNH_1320400 | 277 S | STRSSTHHP | 0.993 | unsp | PKNH_1320400 | 485 S | PKGASKKRQ | 0.996 | unsp | PKNH_1320400 | 491 S | KRQKSGNKE | 0.996 | unsp | PKNH_1320400 | 521 S | TQDDSDNDT | 0.995 | unsp | PKNH_1320400 | 66 S | PEDTSPHSQ | 0.991 | unsp | PKNH_1320400 | 69 S | TSPHSQEEE | 0.998 | unsp |