

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PKNH_1330400 | OTHER | 0.999882 | 0.000071 | 0.000047 |
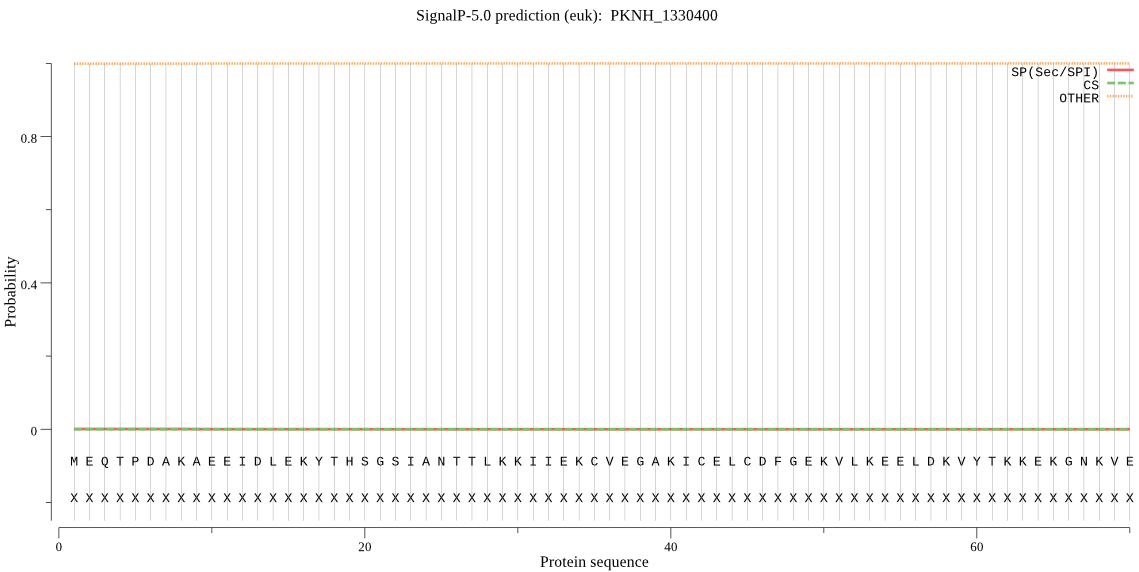
MEQTPDAKAEEIDLEKYTHSGSIANTTLKKIIEKCVEGAKICELCDFGEKVLKEELDKVY TKKEKGNKVEKGISFPVTINVNEVCNNYSPAPSDNEETIKSGDIVKICLGCHIDGHISMV GHTIYVGKENEVIEGPKAEVLKNAHTISQLFLKSLKVGVNASDITKNIQKACEELKCTVI PNCVTYQIKKYILEGSKFISLKENPENKIEDFQIESDDIYIIDVMVTTGDGKIKESDHKT TIYKREVQKNYHLKTNLGRSFINEVNKKFPVLPFHVKHVEDQRAALIGIPEALRHDLIKP YNVYTEKKKEFVSQFKYTVMVKEEGIKQLTGIKCSQLNNCKTVNEIQDETIKAILATSLT AKKKKSKGKPNGETPQDN
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| PKNH_1330400 | 200 S | SKFISLKEN | 0.995 | unsp |