

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
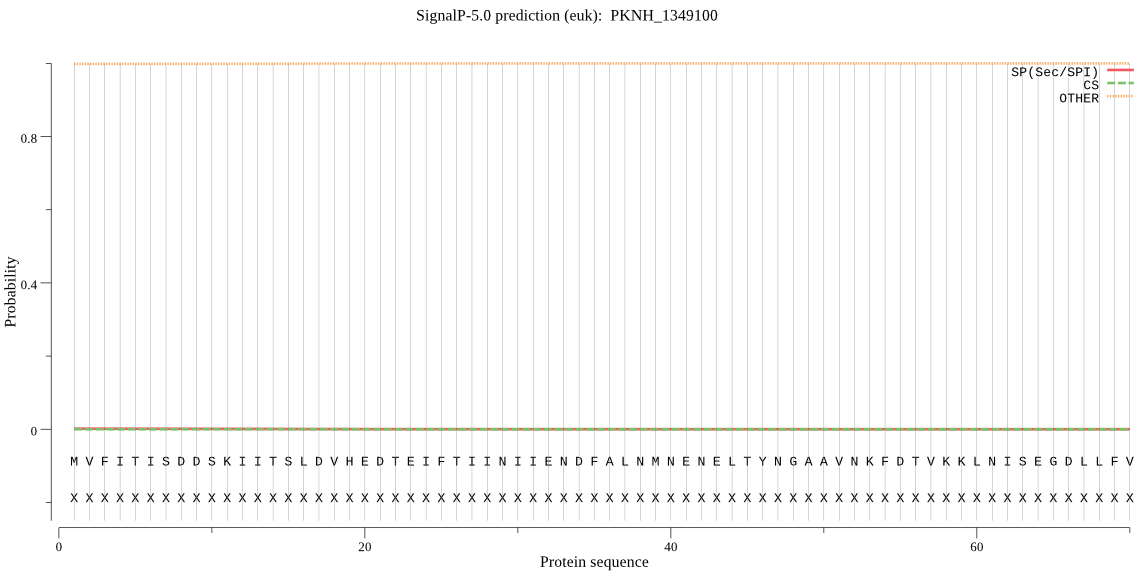
| PKNH_1349100 | OTHER | 0.999864 | 0.000136 | 0.000000 |
MVFITISDDSKIITSLDVHEDTEIFTIINIIENDFALNMNENELTYNGAAVNKFDTVKKL NISEGDLLFVRKKLNLDLIPQGNSSTQAGMTAAAVGAASSSTGAGITPTGASGINSNSFN SAGGVNNATFNALMDHFRTFQENEYIKKEVEILLNLKNDRSRMSVLQLQDKQLYDAITSQ DVEAIKKIVKEKLENEKKEKEREQRMYENALKDPLSEDAQKYIFENIYKNQINSNLALAQ EHFPEAFGVVYMLYIPVEINKNVVHAFVDSGAQSSIMSKQCAEKCNILRLMDTRFTGIAK GVGTRSILGKIHMVDIKIGNYFYAVSLTIIDEYDIDFIFGLDLLRRHQCQIDLKKNALVI EDNEIPFLPEKDIIANSSHSIDFDAMKESTRGV
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| PKNH_1349100 | 63 S | KLNISEGDL | 0.99 | unsp |