

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
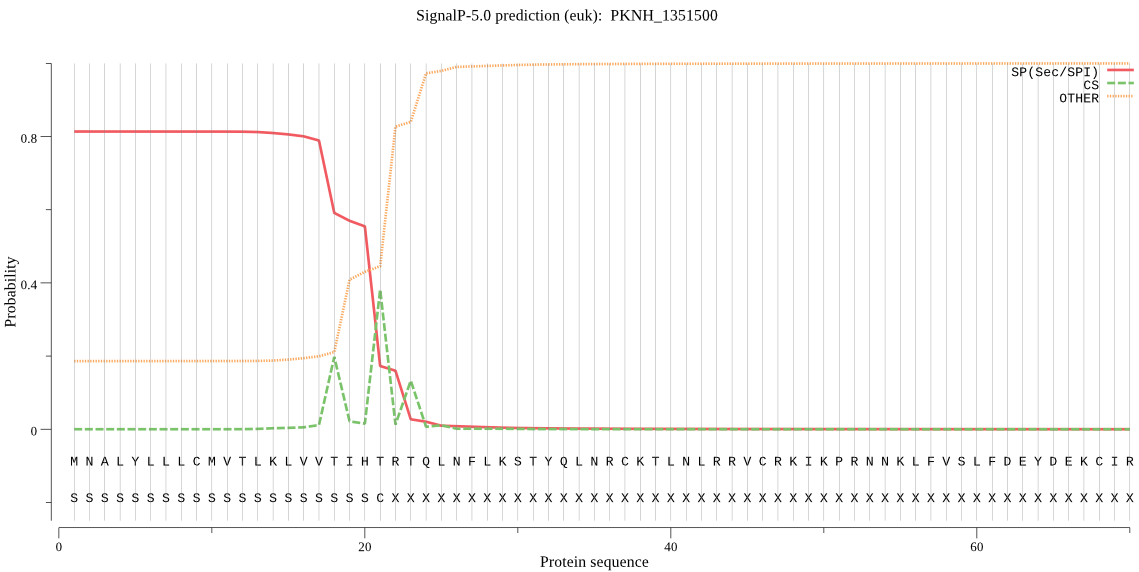
| PKNH_1351500 | SP | 0.132087 | 0.857109 | 0.010803 | CS pos: 21-22. IHT-RT. Pr: 0.6253 |
MNALYLLLCMVTLKLVVTIHTRTQLNFLKSTYQLNRCKTLNLRRVCRKIKPRNNKLFVSL FDEYDEKCIRALIMAREVAKNNNEKEILLKHLLIAIIRIDSNLVKHILNFFNISLTTFLE KLNKEINKKGGETSLQTKQDEQNGSPGGDIPRQAGIEIQETPTNQHHADGRSNDVVEGER TYQSNDQDGEHKLMNTFINKHIQDIEDKINQLKNIDKGQSDISREVYDSDNSVEGSDERD ETTRNLIHDAAKRVLGNHSGDLEDAPDGPPLLDPENGDQLPDELSSDLSKELSRQLPRDL PNGLPNELPDEKADIKFSENCKRALHNAVLEARRKKKMFVNVTDILIALINMAQENENCE FLKYLSELNISVNDLKRELIGYDEENSFAPPTGRTSHSDQRDDDQSSNQADRREENGSLM EAIENRDDNQRMRNVNNEQSNHNFMNNLNNDYLNQTREFKNYEENSFPSNNKLFGSSYVK DCLTDMVHAAYEKGDEHFFGRKKEIKRIIEILGRRKKSNPLLIGESGVGKTAIVEHLSYL ILKDKVPYHLRNCRIYQLNVGNIVAGTKYRGEFEEKMKHLLSNMNKKKKNILFIDEIHVI VGAGSGEGSLDASNLLKPFLSSDNLQCIGTTTFQEYTKYIEADKALRRRFNCVPVKPFSA KETLLLLKKIKYNYEKYHNIYYTNDALKSIVMLTEDYLPTANFPDKAIDILDEAGAYQKI KYEMFMRQRLRDERGRRADAEPRNAESANGDLANGNLTNGEQMISHQVNATETSSDDHPP GKLRTDDNQPAFVHEDRNVEAENYLENMHMKYVTSDVIENIVSKKSSISYIKKNKKEEEK IIKLKEKLNKIIIGQEKVIDILSRYLFKAITNIKDPNKPIGTLLLCGSSGVGKTLCAQVI SKYLFNEDNLIVINMSEYIDKHSVSKLFGSYPGYIGYKEGGELTESVKKKPFSIILFDEI EKAHSEVLHVLLQILDNGMLTDSKGNKVSFKNTFIFMTTNVGSDIITDYFKLYNKNYANL GFKYYLNKKKNKEETDGSSLHKGQLPQGVNSVGNSVVEHPTGLSNNSATDVQINHKRDHE VENPPTQQPSSYELFEEKLRTNEWYEELQPEIEEELKKKFLPEFLNRIDEKIIFRQFLKR DVVNIFQNMIEDLKKRIKKRKNLNLIIEQDVIKYICSDENNIYDMNFGARSIRRALYKYI EDPIASFLISNLCEPNDSIHVSLSNGNKINVQLLKASVDQLVL
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PKNH_1351500 | 396 S | TGRTSHSDQ | 0.998 | unsp | PKNH_1351500 | 396 S | TGRTSHSDQ | 0.998 | unsp | PKNH_1351500 | 396 S | TGRTSHSDQ | 0.998 | unsp | PKNH_1351500 | 659 S | VKPFSAKET | 0.998 | unsp | PKNH_1351500 | 1091 S | QQPSSYELF | 0.993 | unsp | PKNH_1351500 | 223 S | QSDISREVY | 0.991 | unsp | PKNH_1351500 | 232 S | DSDNSVEGS | 0.995 | unsp |