

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PKNH_1401500 | OTHER | 0.999993 | 0.000006 | 0.000002 |
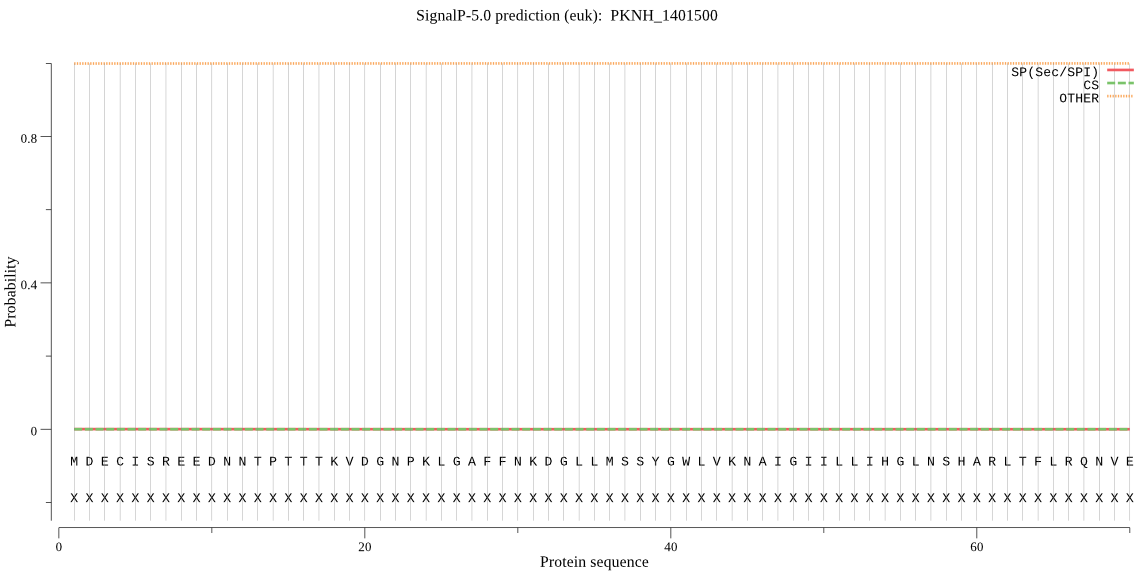
MDECISREEDNNTPTTTKVDGNPKLGAFFNKDGLLMSSYGWLVKNAIGIILLIHGLNSHA RLTFLRQNVEIVDNDKVILKDANNYYVYKDSWIEHFNKNGYSVYALDLEGHGLSDGWGDL SLNINKFDDLVHDVIQYLNIINDDLCLEENEKNSKAICDKAKECTTLCNYDNGSNNDKCK RNVRSNNKRKREKASKGTNNPQSNDKPCSSKEHTNNKCSSLPIYILGQSMGGNIALRTLQ VLEKTQNNGKGRLNIQGCISLSSMICFQKIASPRSYKYKIFYLPFTRLIGSLFPTLRLAT KIGFQKYPYLNQLSNFDNITSKIGLTLKYWCELVKATSNLEKDMRYMPKDIPILFIHSKE DIFCLYRGVVSFFNRLDNDNKELISLDNMEHGLTTEPGNEIVLEKIVNWIKKLKTKKQMT KSLKNK
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PKNH_1401500 | 358 S | LFIHSKEDI | 0.996 | unsp | PKNH_1401500 | 358 S | LFIHSKEDI | 0.996 | unsp | PKNH_1401500 | 358 S | LFIHSKEDI | 0.996 | unsp | PKNH_1401500 | 209 S | DKPCSSKEH | 0.994 | unsp | PKNH_1401500 | 272 S | QKIASPRSY | 0.992 | unsp |