

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
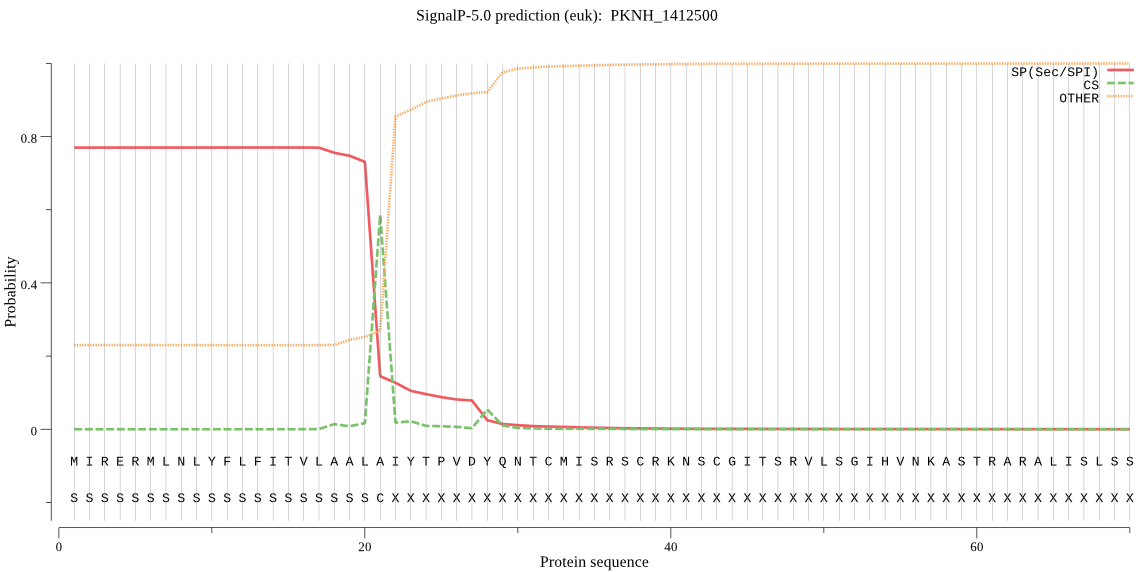
| PKNH_1412500 | SP | 0.139815 | 0.859823 | 0.000362 | CS pos: 21-22. ALA-IY. Pr: 0.5999 |
MIRERMLNLYFLFITVLAALAIYTPVDYQNTCMISRSCRKNSCGITSRVLSGIHVNKAST RARALISLSSLIYQLQLPKLVSLDLFRRDLFTGVNQKGRRSPVPSYIIQSRLMSENIDSG NNNMSATGNQEKKRPVTGDASDAKNPDGSTISAQDKRMKGIDQNEGTSSVSGSTNAAFTN GASSIMEGGENNNNGTGNEGKNEPTIHYRKDYRPSGFIIDNVTLNINIFDNETSVRSTLD MKLSDHYRGEDLIFDGVSLEIKEISIDGNKLMEGEHYKYDKEFLTIYSKFIPKGKFTFGS EVIIHPETNYALTGLYKSKNIIVSQCEATGFRRITFFIDRPDMMAKYDVTVTADKEKYPV LLSNGDKLNEFEIPGGRHGARFNDPYLKPCYLFAVVAGDLKHLSDNYVTKFSKRNVELYV FSEEKYVSKLKWALECLKKAMKFDEDYFGLEYDLSRLNLVAVSDFNVGAMENKGLNIFNA NSLLASKKKSIDFSFERILTVVGHEYFHNYTGNRVTLRDWFQLTLKEGLTVHRENLFSEQ TTKTATFRLDHVDLLRSVQFLEDSSPLAHPIRPESYVSMENFYTTTVYDKGSEVMRMYQT ILGDDYYKKGMDIYIKKNDGGTATCEDFNDAMNEAYKLKKGDKTANLDQYLLWFAQSGTP HVTAEYSYDAGKKEFVIDITQVTHPDPNQKEKKALFIPIRVGFINPHNGKEVIPEVTLEF KKDKEKFIFSNVNEKPIPSLFRGFSAPVYIKDNLTDSERIVLLKYDTDAFVRYNVCVDLY MKQIMKNYQELLQAKAENKQESTEKPKLTPVSEDFISAIKYLMEDPHADAGFKSYIITLP RDRFIINSIRNVDTDVLADTKDFIYKQLGDKLNDLYFQIFKSIQAKADDMTHFEDESYVD FEQLNMRKLRNTLLTLLSKAKYPNMLDHIMEHSKSPYPSNWLASLAVSAYYDKYFDLYEK TYNQSKDDELLLQEWLKTVSRSDRKDIYDIIKKLENEVLKDSKNPNEIRAVYLPFTNNLR YFNDISGKGYKMMADIIMKVDKFNPMVATQLCEPFKLWNKLDMKRQDMMLNEMNRMLSME NISNNLKEYLLRLTNKL
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PKNH_1412500 | 237 S | TSVRSTLDM | 0.99 | unsp | PKNH_1412500 | 237 S | TSVRSTLDM | 0.99 | unsp | PKNH_1412500 | 237 S | TSVRSTLDM | 0.99 | unsp | PKNH_1412500 | 426 Y | SEEKYVSKL | 0.993 | unsp | PKNH_1412500 | 802 S | NKQESTEKP | 0.996 | unsp | PKNH_1412500 | 898 Y | EDESYVDFE | 0.992 | unsp | PKNH_1412500 | 1078 S | NRMLSMENI | 0.995 | unsp | PKNH_1412500 | 101 S | KGRRSPVPS | 0.995 | unsp | PKNH_1412500 | 184 S | NGASSIMEG | 0.995 | unsp |