

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PKNH_1421000 | OTHER | 0.965742 | 0.003779 | 0.030479 |
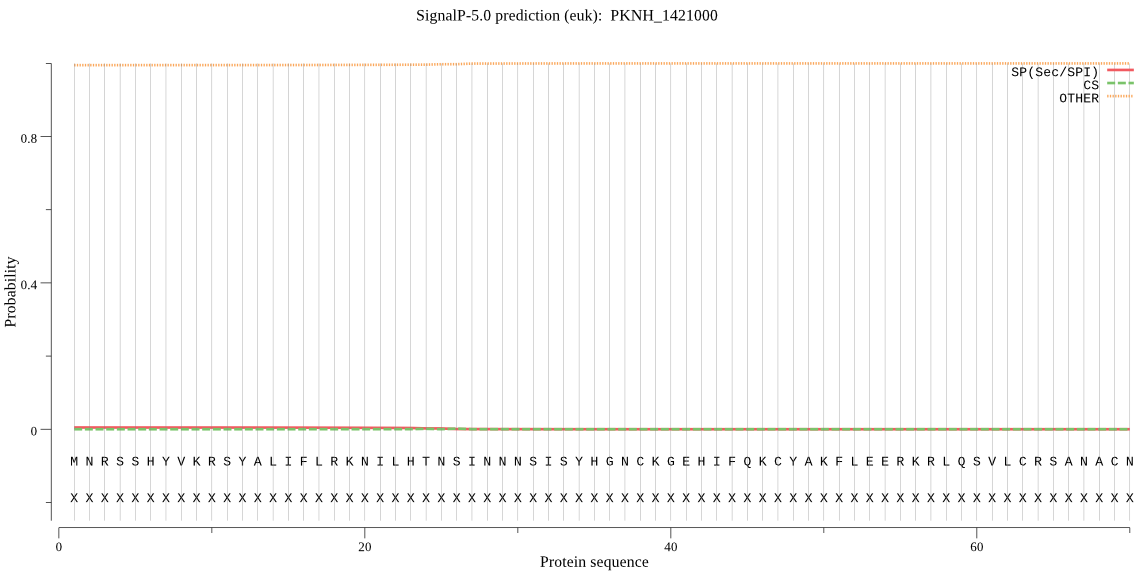
MNRSSHYVKRSYALIFLRKNILHTNSINNNSISYHGNCKGEHIFQKCYAKFLEERKRLQS VLCRSANACNFVLSPRSRGSSLKCYSERKKRGNNKYRTRRRNFFFFNFVKINNPLNFIKK IILSFLLIFGINNYVIDMTLTSGSSMCPLINKNGVILFYVCDDTVRFIHQARSIFLYSCI NLLLRCYALIGSNIEQSYMVILNNKIFSLIEKLKRIMAENKHVYRRGDVILLTSPVNEKK RVCKRIIAIGNDKLFVDNIKAFVHVPKDNVWVEGDNKMDSFDSRNYGFVHMDLIIGRVIF LLDPFINFRFISNRTS
| ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|
| PKNH_1421000 | 81 S | SRGSSLKCY | 0.997 | unsp |