

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
| PKNH_1428400 | OTHER | 0.991600 | 0.004961 | 0.003439 |
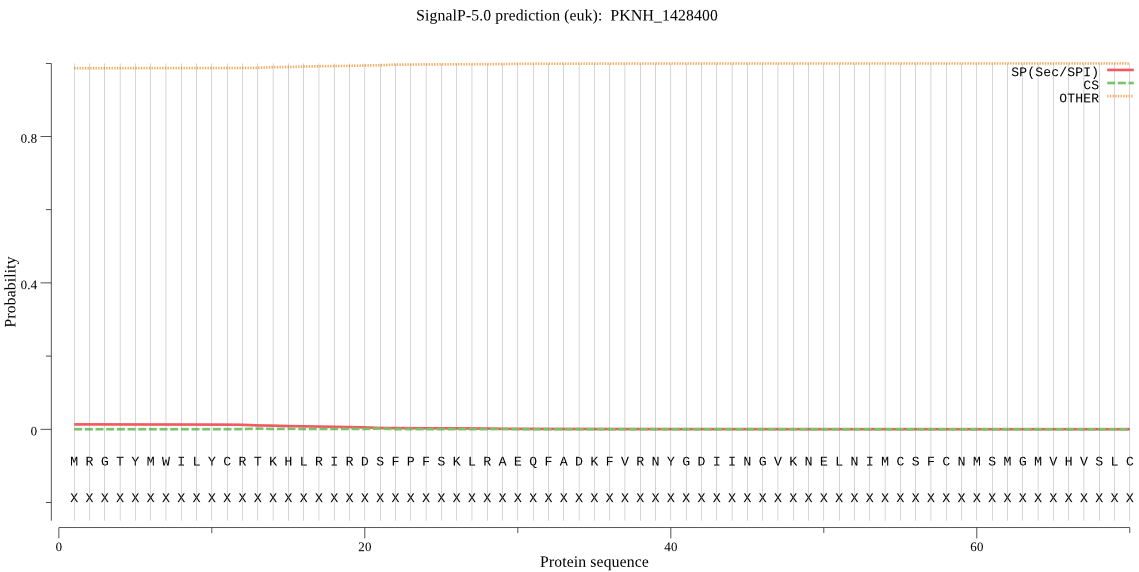
MRGTYMWILYCRTKHLRIRDSFPFSKLRAEQFADKFVRNYGDIINGVKNELNIMCSFCNM SMGMVHVSLCESAIGSPITPVDGCDCTFEHKPEHRLSHGAHDAPSENDQLEQNTQPEKKG TSKLATYLIIYYVKEYVKYVAHSLLHFCFNLCDTIFRTLFVLPKSYLEFPKQLDYYSSIS DNLLTSFVTIYKIYKQNDSSEFRFKLDQLAFLGSGFIYDKRGYVLTAAHNITNTEGTFVI KNEDNFYVATVAGLHRESDICVMKIISEEQFNHIPLNPSRELLKPGETVITYGQIQNFDK GTCSIGVVNQPRQTFTKFENFSEKEKTCLYPFIQISNPINKGMSGSPLIDKHGNLVGMIQ KKIDNYGLALPVNILKNITTHLQEEGTYKEPFLGIVLREKEQSISAVYKNCKNEELKIQD VLVNSPADVAGITKGDIMLRINNTKIKNICDVHEILNSTSDGYVTVDIIRHGKNKKIKVK M
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PKNH_1428400 | 322 S | FENFSEKEK | 0.994 | unsp | PKNH_1428400 | 322 S | FENFSEKEK | 0.994 | unsp | PKNH_1428400 | 322 S | FENFSEKEK | 0.994 | unsp | PKNH_1428400 | 97 S | EHRLSHGAH | 0.996 | unsp | PKNH_1428400 | 105 S | HDAPSENDQ | 0.992 | unsp |