

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
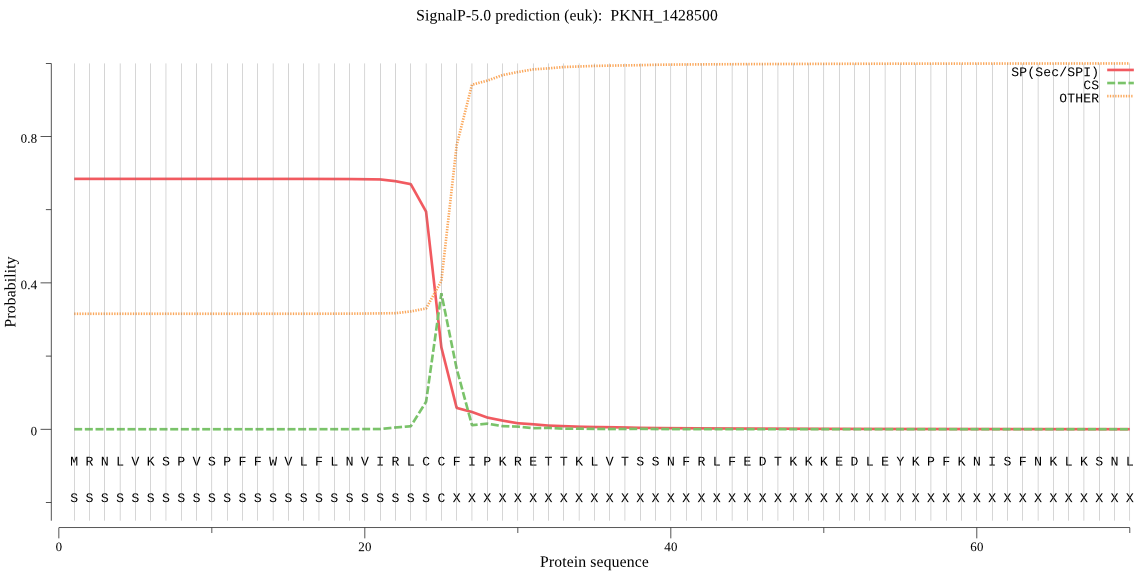
| PKNH_1428500 | SP | 0.215443 | 0.677533 | 0.107024 | CS pos: 25-26. LCC-FI. Pr: 0.4589 |
MRNLVKSPVSPFFWVLFLNVIRLCCFIPKRETTKLVTSSNFRLFEDTKKKEDLEYKPFKN ISFNKLKSNLKNFLVVNKKFSISDVKQVSFLGKFYNYEKIENYGVNEICILGRSNVGKST FLRNFIKYLINVNEHANVKVSKNSGCTRSINLYSFENGKKKKLFILTDMPGFGYAAGIGK KKMEFLRKNLEDYIFLRNQICLFFVLIDMSVDIQKIDVSIVDAIKKTNIPFRVICTKSDK FSTKADERLEAIKNFYQLEKIPIHISKFSTHNYINIFKEIQHHCNLDIP
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|
| PKNH_1428500 | 81 S | NKKFSISDV | 0.992 | unsp | PKNH_1428500 | 242 S | SDKFSTKAD | 0.997 | unsp |