

| _ID | Prediction | OTHER | SP | mTP | CS_Position | |
|---|---|---|---|---|---|---|
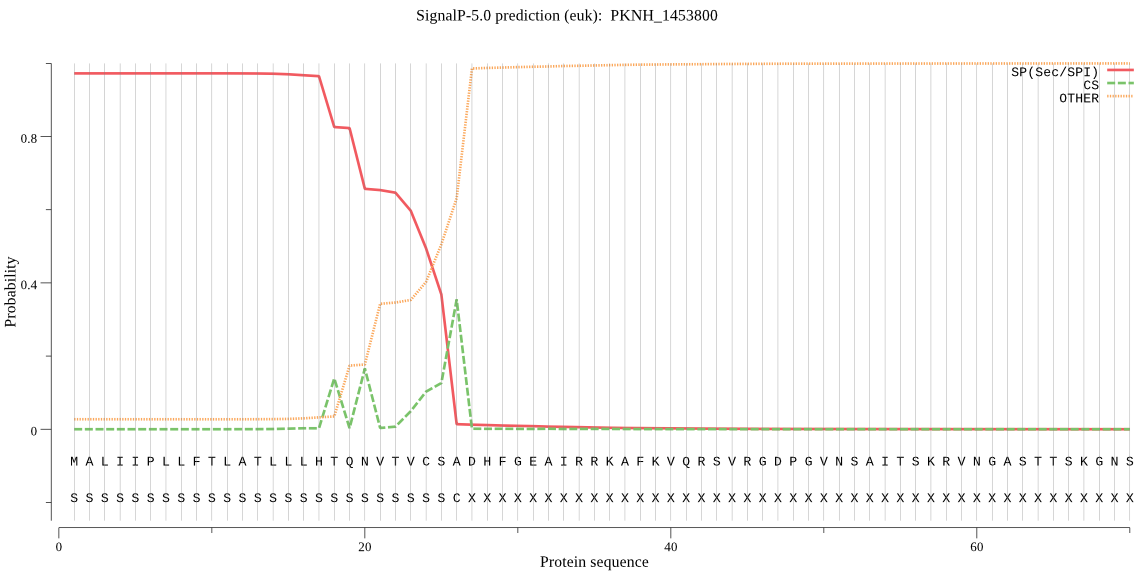
| PKNH_1453800 | SP | 0.004930 | 0.994763 | 0.000307 | CS pos: 26-27. CSA-DH. Pr: 0.2757 |
MALIIPLLFTLATLLLHTQNVTVCSADHFGEAIRRKAFKVQRSVRGDPGVNSAITSKRVN GASTTSKGNSLLEAKKREQDYFTLKINEQNFRWSLPLLMGSKKTPLELGIVTSTPITALY CSYNAKPNEEMNNIKYEVNASEGVKYISCKSRYCTAIGQGNTCAPIEHFFKMIHDFGLRK KNCTNRFCTYINDINFLNVNTKLDKRNMSVCSFSSNLGSEQVEGFYFRDSFYLYDTVKFD YKHFGCVTQSGVLTFNNVISGFIGLAYNRADAIANKKESSSILHTLVQKSVSKKNIFGLC FVEGGGFATFGGINNEALRKVLPVSKLQMGFQHLGGEDPQATSHEIVWLAYSDTSKSTYS LLLKEVNMVSTSNRVENAIGRVAVIDSYNYFLSFPAEITAKLKTAIYNSCVGNDNKCSNI INKGVFTLKNGGVDDFPTVELVFDDGKVLIHPKDYLIHEGDGVYRVLINSEETLKLGIPF FLNKYLTFDNENGKIGVGPSDCTFEMKGVSPGVDLSATEADSNDDPEDKDFTIEDFFQEN KLMILALTSLSVVGAIVGGVFFFG
| ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | ID | Site | Peptide | Score | Method | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PKNH_1453800 | 292 S | QKSVSKKNI | 0.994 | unsp | PKNH_1453800 | 292 S | QKSVSKKNI | 0.994 | unsp | PKNH_1453800 | 292 S | QKSVSKKNI | 0.994 | unsp | PKNH_1453800 | 522 S | TEADSNDDP | 0.996 | unsp | PKNH_1453800 | 43 S | KVQRSVRGD | 0.992 | unsp | PKNH_1453800 | 70 S | SKGNSLLEA | 0.992 | unsp |